 |
Top : Molecular Biology : DNA : DNA Extraction & Purification : DNA Extraction from Plants : Development of a rapid and efficient method for DNA extraction from plants containing high amount of
Development of a rapid and efficient method for DNA extraction from plants containing high amount of | |
| Author: H. Momeni, B. Shiran, M. Kohgard,M. Khoddambashi | |
| Affiliation: Department of Agronomy and Plant Breeding, Faculty of Agriculture, Shahrekord University, Shahrekord, Iran.
*Author for correspondence: Hassan momeni, Department of Agronomy and Plant Breeding, Faculty of Agriculture, Shahrekord University, Shahrekord, P.O.Box 115, Iran, e-mail: Hassan_momeni17@yahoo.com Fax number: +98 3814424412 | |
| Source: Protocol Online | |
| Date Added: Tue Jan 18 2011 | |
| Date Modified: Sat Feb 26 2011 | |
| Abstract: Extraction was repeated several times using chloroform: isoamylalchohol to obtain more purified DNA. Average yield of DNA extracted with this procedure was 700-1500 ng/microL from 100 mg fresh leaf tissue. Extracted DNA showed complete digestion with restriction endonuclease and successful reproducibility through PCR amplification. This method, which is relatively inexpensive, could be efficiently applied for other species of different families that contain polyphenols and secondary methabolite is cumbersome. | |
Introduction
Fascinating Fritillaria imperialis which also is called ‘‘Tears of Mary’’,
because of great drops of nectar at the petal base [5], is a perennial plant
with high medicinal and ornamental importance and is the most famous species in
the Fritillaria genus [1]. Iranian native species of F. imperialis have been
reported as endangered plant species [5]. Subsequently in order to preserve the
species, molecular assays have to be carried out and making a good understood
about this germplasm. At the other hand existence of polyphenols and secondary
methabolite is cumbersome in PCR-based methods.These studies require analysis of
large number of samples, thus a DNA extraction method, which is fast,
inexpensive and yields high quality DNA, is desired. Several methods for
extraction of genomic DNA from plant tissues have been carried out and
demonstrated so far. Also this subject has been developed by improvement of
science [2,3,4]. We have now modified the CTAB method for rapid isolation of DNA
from plant. The DNA is suitable for ISSR analysis and other PCR-based
applications.
Materials and Methods
Sample collection
Fresh bract leaves were collected from different region in Iran and were took to laboratory. For each sample, three parcels were made and reserved in -70 refrigerator until DNA extraction.
Standard chemicals
This method uses standard chemicals that can be obtained from any major
supplier; we used chemicals supplied by Sigma Co. as follow:
- Extraction buffer contains: NaCl (1.4 M); Tris-HC1 (100 mM), pH 8.0; EDTA(20 mM), pH 8.0;CTAB powder per 100 ml and β–Mercaptoethanol (1%).
- Chloroform : Isoamylalcohol (24:1); RNase A (10 mg/ml); Isopropanol and ethanol (90%).
Procedure
- Preheat CTAB buffer with 1% β -Mercaptoethanol (added just before buffer use) in water bath at 65˚C.
- Grind plant tissue with micro-pestle in liquid nitrogen until it is turned into fine powder and put 100 mg of powdered leaf material in tube.
- Add 750 µl extraction buffer (CTAB+ β-Mercaptoethanol) per tube, mix well by inversion and incubate in water bath 3 min at 65˚C.
- Add 750 µl Chloroform : Isoamylalcohol (24:1), mix well by inversion 2-3 min and centrifuge 10 min 13000 rpm.
- Transfer the top phase to a new tube (approx. 600 µl).
- Repeat Chloroform : Isoamylalcohol step.
- Add 2/3 of top phase volume (approx. 450 µl) of cold Isopropanol (kept at -20˚C) to precipitate DNA and mix by inversion.
- Centrifuge 5 min 13000 rpm, pure of supernatant, wash DNA pellet by adding 500 µl cold 70% ethanol (kept at -20˚C) and mix by inversion.
- Centrifuge 5 min 13000 rpm and carefully remove ethanol with pipette (since pellet is easily loose at this stage).
- Dry pellet at room temperature overnight or in speed vac (some scientists prefer former as less invasive, but this way is also OK).
- Resuspend DNA in 40 µl H2O (or more H2O can be added later after quantification DNA if necessary).
- Add RNase A to the final concentration of 1 µg/µl and incubate 30 min at 37˚C.
- Store isolated DNA at -20˚C.
DNA can be quantified and diluted to a working concentration at this point or simply use 1 µl per PCR reaction. We expect that the yield of this procedure be 100 to 300 ng/µl, DNA. Using the above method, high quality DNA samples from a plant population were extracted for molecular genetic studies.
Results
The quality and quantity of extracted genomic DNA were controlled. High quality DNA was obtained using our method. All of the samples were able to profiling for ISSR and other PCR applications. Agarose gel 1.5% was used for quality control of genomic DNA. Extracted genomic DNA by our method on Agarose gel 0.8%, ISSR amplification and Digestion of DNA with EcoR1 and Xbal restriction enzymes are given in Figure 1. The yields of the DNA samples ranged from 700 to 1500 ng/μl from 100 mg fresh leaf tissue that was stored at -70 ºC for one year. This amount of DNA is enough to conduct 200 to 300 PCR reactions. It is easily done and the gDNA is pure and not degraded.
Discussion
Extracted genomic DNA from different biological samples is used widely in molecular genetics assays. In our method, genomic DNA can be extracted in the least time and with high quality and quantity by using simple materials and equipments.
Not only was high quality DNA extracted from leaf tissue that was stored at 4ºC, this method also worked well for extracting DNA from the other wild plant samples that were stored at -20ºC or -80ºC. In a workday, one person can complete DNA isolation from more than 50 plant samples using this method. This method has been routinely used to extract DNA from F. imperialis leaves for PCR based applications in our laboratory but it can be used for other species of different families that contain polyphenols and secondary methabolite
This method has several advantages such as economical spending, no need to the specialized and expensive equipments, spending little time, no need to the experimented and experienced staff and more important, DNA extraction from plant leaves stored at usual fridges for long time. In this method, genomic DNA with high quality and quantity can be acquired for other species that contain polyphenols and secondary methabolite is cumbersome. Time of extraction of genomic DNA in our method is less than three hours.
Acknowledgments
The authors offer grateful thanks to Shahrekord University for financial assistance; as well as to the staff of Natural Resources Organization for their approval to Sampling.
References
- Alp, S., Arsalan, N. and Koyunku M. (2009). established forms of Fritillaria imperialis L. –A naturally growing species in Turkey. Pakistan Journal Botany. 41(4): 1573-1576.
- Dnyaneshwar, W., Preeti, C., Kalpana, J. and Bhushan, P. (2003). DNA isolation from fresh and dry plant samples with highly acidic tissue extracts. Plant Molecular Biology Reporter 21: 467a–467f.
- Guillemaut, P and Mardchal-Drouard L.1992. Isolation of plant DNA: A fast, inexpensive and reliable method. Plant Molecular Biology Reporter. Volume10(1): 60-65.
- Lin, R.C., Ding, Z.S., Li, L.B and Kuang T.Y . (2001). A Rapid and efficient DNA minipreparation suitable for screening transgenic plants. Plant Molecular Biology Reporter. 19: 379a–379e.
- Mohammadi-dehcheshmeh M, Khalighi A, Naderi R, Ebrahimie E, Sardari M (2007) Indirect somatic embryogenesis from petal explant of endangered wild population of Fritillaria imperialis. Pakistan Journal Biology Science 10:1875– 1879.
- Ribeiro, R.A. and Lovato M.B. (2007). Comparative analysis of different DNA extraction protocols in fresh and herbarium specimens of the genus Dalbergia. Genetics and Molecular Research 6 (1): 173-187.
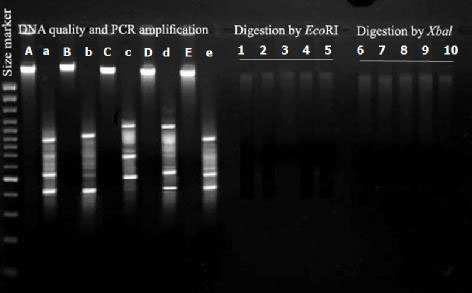
Figure 1. Genomic DNA extracted from fresh bract leaves of the different Tears
of Mary Samples using the rapid isolation method on agarose gel; Lanes A, B, C,
D and E. ISSR- PCR analysis of DNA samples isolated by the rapid isolation
method; Lanes a, b, c, d, e and f. DNA digestion by EcoRI restriction enzyme;
Lances 1 to 5. DNA digestion by Xbal restriction enzyme; Lances 6 to 10.