 |
Top : Cell Biology : Cell Culture : General Procedures : Cell Counting and Viability Assay : Cell Counting with the Sedgewick-Rafter Chamber and Whipple Micrometer Disc
Cell Counting with the Sedgewick-Rafter Chamber and Whipple Micrometer Disc | |
| Author: Stephen M. Gittleson and Mandanna Ganapathy | |
| Affiliation: Fairleigh Dickinson University, School of Natural Sciences 1000 River Road, Teaneck, NJ 07666. E-mail: gittleso@fdu.edu | |
| Source: Protocol Online | |
| Date Added: Tue Mar 08 2011 | |
| Date Modified: Sat Apr 09 2011 | |
| Abstract: A step by step protocol to standardize the counting of cells using the Sedgewick-Rafter Chamber and Whipple Micrometer Disc. Statistically significant counts of protists in cultures of millions/ml may be obtained by directly counting less than 0.1% . | |
Summary. A step by step protocol to standardize the counting of cells using the Sedgewick-Rafter Chamber and Whipple Micrometer Disc. Statistically significant counts of protists in cultures of millions/ml may be obtained by directly counting less than 0.1% .
Abbreviations: SRC-Sedgewick-Rafter Chamber, WMD-Whipple Micrometer Disc, P. papillata-Polytomella papillata, CC-Calibration Constant, Confidence Interval (CI),
IntroductionWe identify and control sources of error such as those related to geometry of the SRC, sample volume, method of dispensing the sample and settling behavior of the preserved organisms. The The flagellate P. papillata is referred to here but this cell counting technique should be applicable to other free-living protozoa, algae, and phytoplankton.
ProcedureThe SRC without grid with cover glasses is available from Wildlife Supply Company, 95 Botsford Place, Buffalo NY 14216 (www.wildco.com ). The WMD is available from Max Levy Autograph, Inc., 220 W. Roberts Ave, Philadelphia PA 19144 (www.maxlevy.com ). Each step of this protocol is supplemented with comments in an attempt to minimize confusion over ‘obvious’ manipulations that may influence the repeatability and accuracy of this technique. Sample data, calculations and statistics appear in the results and discussion.
- Calibration of the SRC with WMD. The WMD is installed in the eyepiece of the microscope so that the WMD grid appears in the field of view through the microscope as illustrated in this figure:
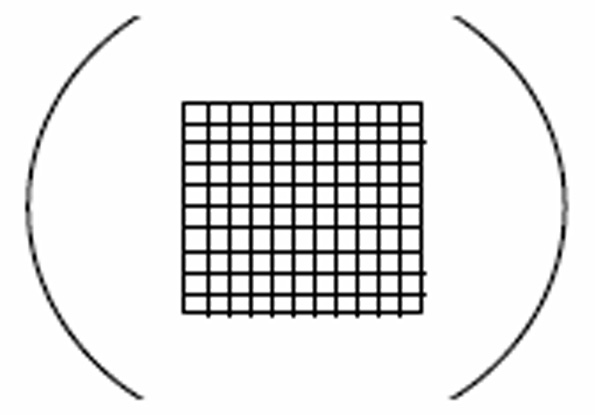
Comments for step 1: This calibration procedure establishes the relationship of the number of organisms counted under the WMD grid to the total number of organisms in the sample within the SRC. A 10X ocular and 10X objective is used here, but other lens magnifications appropriate for counting organisms of different sizes, can be selected. A separate calibration is necessary for each different arrangement of lenses and for different microscopes . The WMD contains a grid composed of 100 equivalent squares. Dimensions of the squares are measured with a slide micrometer. In our arrangement, the side of each square measures 0.16 mm. Thus, each square has an area of 0.0256 mm2. . Since the SRC measures 20 mm by 50 mm it has an area of 1000 mm2 . The portion of the SRC covered by one square of the WMD grid is 1000 mm2 divided by 0.0256 mm2 . Therefore, the particular CC for our setup is 39,062 which is the number of WMD squares needed to cover the entire SRC.
- Swirl the culture manually or better with a mechanical agitator, such as the Vortex Genie ( www.scientificindustries.com/genie2.html ) to randomize the organisms.
- Withdraw a 1.0 ml sample from the culture and dispense it into a test tube. Repeat Step 2 and then withdraw another 1.0 ml sample from the culture and dispense it into a second test
tube.
Comments: Using a 5 ml serological pipette, Pyrex brand disposable (Corning 7077-5N) calibrated in 0.1 ml graduations delivery to the tip. This pipette is the type graduated to the tip and has a permanently marked band or bands near the top end to indicate that a small amount remaining in the tip after free delivery has ceased, must be blown out to obtain total rated capacity. The Bulb-Type Safety Pipet Filler (Fisher Scientific Cat.No. 13-681-50) is useful for safe, easy and repeatable pipetting. We used a 16 X 150 mm disposable glass tube. - Add 3.0 ml of a 10% formaldehyde solution to each test tube containing a 1.0 ml sample from the culture (from step 3).
Comments: It is quite possible that another type of preservative will work better with your organism. For example, experiment to determine that your organisms remain intact and do not clump during the cell counting procedure. - Swirl the preserved organisms in the first test tube (from step 4) manually or better with a mechanical agitator and withdraw a 1.2 ml sample of these preserved organisms.
- Dispense the entire 1.2 ml sample of preserved organisms into the SRC and place the cover glass over the chamber avoiding air bubbles.
Comments: The best random distribution of preserved organisms in the SRC is accomplished by dispensing 1.2 ml of preserved organisms from the pipette using the pattern shown in this figure.
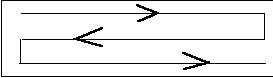
Placement of the cover glass limits the volume to 1 ml.
- Allow the preserved organisms in the SRC to settle for 5 min before counting.
Comments: Other species may require a different settling time so before counting verify that all of the preserved organisms are at the bottom. The count should be taken as soon as possible. Extended exposure to the formaldehyde may cause clumping or other changes which influence the count. - At each of two randomly selected locations in the SRC, count the number of organisms in 50 WMD squares. Where organisms lie on the lines of a square, include in the count, only those organisms on
the right side and bottom side.
Comments for step 8: The locations within the SRC where counts are made are selected at random by using a set of random numbers selected at random Randomly selected locations on the edge are rejected. Each set of random numbers is recorded on a slip of paper and placed in a container from which three slips are blindly removed. After using these three sets of random coordinates for counting at three locations in one sample the slips of paper are returned to the container holding all slips of paper. Slips of paper with the location coordinates are randomly selected for counting in each sample. The lower left corner of the SRC is taken as the origin (0,0) from which the random spots are located. - Repeat steps 5 through 8 to count organisms from the second test tube (the parallel sample).
Comments: Parallel samples are compared to monitor the occurrence of technical errors. The means may be analyzed using a test of significance such as the t-test.
The final calculation for cell count in the original culture is carried out using the following formula: C = (DF) (NC) (CC) / S
C-cells/ml, DF-dilution factor, NC-cells counted, CC-calibration constant, S-number of WMD squares containing NC
For example, using the mean NC between counts 1 and 2 in Tables 1 and 2 C = (4) (314) (39,062) / 50 C = 981,237
Thus, statistically significant counts of organism numbers in cultures of millions/ml may be obtained by directly counting less than 0.1%.