Identification of the cut and uncut plasmid on gel - (Jun/27/2005 )
hi to everyone.
would u plz tell me how we will differentiate between the gel electrophoresis of cut and uncut plasmid( in reference to pBluescript).
i will be highly obliged.
if possible can u show me the photo.
Here is a picture.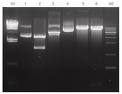
M1 : Lambda Hind III marker,
lane 1: pGEM-T(uncut, insert 1kb),
lane 2 : pGEM-T(EcoRⅠ),
lane 3 : pGEM-T(SphⅠ),
lane 4 : pUWL201(6.40 kb) ,
lane 5 : pUWL201(EcoRⅠ),
lane 6 : pUWL201(XbaⅠ)
M2 : 1 kb ladder marker.
would u plz tell me how we will differentiate between the gel electrophoresis of cut and uncut plasmid( in reference to pBluescript).
i will be highly obliged.
if possible can u show me the photo.
As a rule, the uncut plasmid is supercoiled, so it will run faster that the linearized (cut) plasmid.
Just run your digested sample next to the uncut plasmid you can identify it.
hi veteran,
really feeling very nice to see your warm response.
actually veteran i have one more problem, that my vector is 2.9kb. whenever i isolate my plsmid by mini prep(DH5, E.Coli strain) , i get only one band of around 2.3kbp.
why dont around 2.9 kb. yet the transformtion is very normal by CaCl2 method.
i used the alkline lysis method for plasmid isolation.
if u have any information regarding this, then plz guide me , i will be be very thank ful to u.
I would guess that supercoiled pBluescript would run at about 2.3kB. Linearize it with NotI and run it next to undigested. Compare the migration pattern you see to your marker bands.
