Linearizing pSUPER.retro.puro - (Aug/03/2006 )
I have been trying for the past month to clone a shRNA oligo pair into the pSUPER.retro.puro BglII/HindIII sites. However, I get an empty vector background that makes it impossible to find a colony that contains the insert. I believe it is because the two sites are so close together and I'm not getting complete cutting. I have overdigested plasmid already cut with HindIII with 30X extra BglII and still get the background. I have also tried CIPing the vector to no avail. Does anyone have any other suggestions? Thank you.
This link gives all the details which you might already have. We have cloned shRNA in the same vector between BglII and HindIII. As suggested in the manual we did sequential digestion:
First digested with NEB HindIII for 2Hrs at 37C in NEB Buffer2. Then added 1ul NEB2buffer and BglII. (without any heat inactivation). Continued digestion for further 2Hrs and followed by dephosphorylation by Alkaline phosphatase for 30' at 37C. Approximately 1:4 of V:I was ligated. COnfirmation was done first by digesting with BglII(upon successful ligation BglII site closes) and uncut clones were further cinfirmed by EcoRI-HindII digestion. Successful clone must give a band at~281bp and negative at 227bp.(Check the attached picture)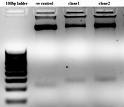
i digest in neb2 by hind III ethanol precipitate, and digest by Bgl2 in NEB3. Then CIP treatment and go for ligation.
I prefer digesting by Eco/Hind in EcoRI buffer + BSA for checking succesful ligation
