Advice on sonication (c2c12) - (Apr/19/2012 )
Hi everyone,
I am still having a few issues with sonication and really hope someone can help me out this time ![]()
I am sonicating C2C12 cells with a Sonics Vibracell using a 3mm stepped microtip, in approx 500ul. I'm using 25% amplitude, 20 sec on/30 sec off. The problem is no matter how much I sonicate, I can't seem to shear the chromatin any smaller than 5kb. I've tried different crosslinking times, different power settings, etc and nothing seems to make a difference.
For example (see attached pic):
Lane 1 - 100bp marker
Lane 2 - 15' crosslinking at RT, 15x20s bursts
Lane 3 - 15' crosslinking at RT, 20x20s bursts
Lane 4 - 30' crosslinking at RT, 15x20s bursts
Lane 5 - 30' crosslinking at RT, 20x20s bursts
Lane 6 - 15' crosslinking at 37C, 15x20s bursts
Lane 7 - 15' crosslinking at 37C, 20x20s bursts
Lane 8 - 1kb marker
All samples have been RC, PNK and RnaseA treated. The reason for the crosslinking times is that I was originally doing 30 minutes at 37 degrees (someone else's protocol) and thought this may have been the problem, but cutting the time and temperature back doesn't make a difference.
The other odd thing is that if you look closely, you can see the nucleosomal ladder (I have not seen this happen to anyone else with sonication) and it seems that I am shearing most material down to a single nucleosome.
Any suggestions would be greatly appreciated, I have been trying to figure this out for months!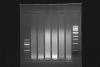
So, are you saying that you can't get the top or your smear of DNA to be smaller than 5kb. That's what the gel appears to suggest. I don't know if you'll be able to get any better than this and would say you're ready to try ChIP. Just use the least intensive shearing conditions that still gives you this size range, as over shearing can denature proteins and decrease your ChIP signal.
maybe pipette your sample between every couple of sonications..............or you could try lowering your sample volume size. How many cells/ml are there?
also, fifteen 20sec pulses seems a lot to me.......
@KPDE - Yes, that is what I'm saying. I've seen pictures of chromatin size that other people seem to get, and it's usually a very tight range that you can see getting smaller and smaller with more sonicating rounds. I tried my same conditions with a completely different cell line (MDA), and got a similar result in that the larger stuff was still there, but then with the MDAs I could also see the differences in chromatin size between rounds (getting smaller and range getting tighter). So I'm now assuming that it is just my cell type, and you're probably right that I can't get any better than what I have. Also, when I sonicate the C2C12 cells without any crosslinking, I get a very nice tight smear that decreases in size with more sonication rounds ![]()
Ultimately I want to do ChIPSeq, so I need a size range of 100-300bp approx. Do you think this is going to be a problem based on how my DNA is shearing?
@chabraha - I am already giving my sample a good mix during sonicating, but thanks for the tip ![]() I am sonicating approx 15x10^6 cells in 500ul.
I am sonicating approx 15x10^6 cells in 500ul.
Here is the MDA cells (top row, 5x20s bursts, 10x20s, 15x20s etc)
and uncrosslinked C2C12 cells (5x20, 10x20 and 15x20).
Just to show what I was trying to say.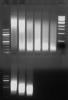
Yeah, cells and tissue are a lot easier to sonicate without crosslinking. Unfortunately, if you don't crosslink and you're looking for TFs or most other non-histone proteins, they're long gone by the time you want to pull them down. Histones, on the other hand, will still be there.
Are you crosslinking C2C12s after they for myotubes? This could certainly give you a harder time with sonication. Last month I was trying to do ChIP on skeletal muscle and that was a pain. I gave up on trying to get any significant amount of small fragments.
I think you should be fine for ChIP-seq since you have a significant amount of material in the 100-300 range. Also, running your input control in parallel will remove any bias for regions that are more easily sonicated. You can at least run ChIP and see how much DNA you're able to get using pico-green or something similar, and if needed, you can pool more than one ChIP of the same type to get more DNA.
Just an update: I think I was over cross-linking due to having sub confluent cells (~50%). This time I harvested cells that were 100% confluent, with the same % formaldehyde and same crosslinking time, and this is my result:
20x20s bursts, and 30x20s bursts
Very happy with how it has sheared this time ![]()

Actually just got the Chip result: the more sonicated sample failed while the other one had ~6-fold enrichment ![]()
Sorry that happened to you. I've had a similar experience, either good shearing or good pulldown. It may be a matter of heating. If you're using a microtip there's not much you can do for the heating problem other than to use shorter pulses with a rest on ice in between, and to leave the tube in an ice/water bath during sonication. It might be worth switching to 10 sec bursts with a 1min rest in ice/water bath in between (still keeping the same total sonication time).
Your lysis buffer is the main cause of problem.
Use this Chip Protocol
https://docs.google.com/open?id=0B15muZJPLjJgbm81UG11X2pUbGlQb3llU2lzampldw
The above protocol is a bit little changed of
Farnham Lab MicroChIP Protocol
https://farnham.genomecenter.ucdavis.edu/pdf/FarnhamLab_MicroChIP.pdf
-----
Babak Memari
