ChIP assay - Foaming in sonication and other woes - (Jan/27/2005 )
Hi all! I was wondering if anyone out there has got any trick to avoid foaming during sonication in the SDS lysis buffer for ChIP assays. Supossedly, placing the tip of the sonicator probe 13 mm below the surface of the liquid should avoid it, but is just not working on my hands. After 10 sec on the first sonication pulse a get lots of foaming, but I like to do longer pulses (20 sec) and foaming is supposed to denature the sample. For how long do you sonicate, and how many pulses? I'm using a Branson 250 sonicator.
Also, which type of vessel do you use for sonication? I use 2 ml eppendorf tubes, but the instruction manual of my sonicator recomends metal or glass vessels, better than plastic ones, because of heating considerations.
And last but not least, have you noticed differences in the sonication quality when using different cell lines? For some reason, I get really small fragments with 3T3 cells but not with 10T/2, and both of them are fibroblast cell lines. I don't change the conditions and sometimes I perform the experiments in parallel ![]()
Any thoughts or suggestions will be very welcome. Thanx!
It is tricky to avoid foaming during sonication. I have tried both 1.5 ml (conival bottom) and 2 ml (round bottom) tubes and found 1.5 ml tubes are less likely to produce foam. In addition, according to this protocol, conical bottom improves sonication efficiency.
To avoid foaming, dip the probe all the way to the bottom of the tube.
To avoid heating, hold the tube in ice water during sonication.
Hope that is useful.
Thanx alot for your answer, pcrman. I'm surprised to see that in the protocol you attach the sonication step is performed in RIPA buffer instead of the more astringent 1%SDS buffer. I've tried before to sonicate the sample in RIPA and the results were dismal, 10 kb DNA fragments. Maybe is because I used larger volumes (2-3 ml) corresponding to the final volume of the IP sample? I see that in this protocol the use smaller volumes and then dilute after sonication. I'll try that. Thanx again!
If you want to avoid foaming during sonication, cup horns are the way to go. They utilize indirect probe sonication, and are able to sonicate sealed vessels. Another advantage is that you can do multiple samples at once. The following link gives a better description:
https://www.misonix.com/Products/index.cfm?...6&div=11&cat=21
Contact Misonix and they will be able to provide lab procedures for DNA shearing using the cup horn.
Hi Badcell,
We use a Branson Sonifier B-30. I had real problems with foaming initially and I came to realise that the chromatin is not shearing properly when it foams up. We were using maximum settings on the sonifier with no luck.
So I tried lowering the settings with the same pulse times. The settings I used and were sucessful are a duty cycle of 30% and power setting to 3. I performed 15 continuous pulses for ten seconds each in 1.5 eppy tubes set in an ice bath to reduce the heat from sonication. We resuspend and sonicate our chromatin in RIPA buffer also.
I would suggest lowering the settings of the sonicator, that certainly stopped the foaming issue and still sheared the chromatin quite nicely.
The cells we work with are CHO hybrids but I am sure different cell lines behave differently. You could try optimising formaldehyde fixation times as I would think cell lines would behave differently to the fixation. (for our cells, 1% formaldehyde in media for 10 minutes exactly is suffice)
hope this helps!
Nick
Thanks all of you for your suggestions, they really help!
Nick, I recently tried something similar to what you said: I fixed the cells using a lower concentration of formaldehyde than usual (0.5% for 10 min) and the shearing seemed to improve. I'll try your suggestion to decrease the power of the sonicator and increase the number of pulses.
Sonix, I thought water bath sonicators are not fit for shearing the DNA for ChIP assays, 'coz they're not powerful enough. I checked the webpage you indicated and it says that this cup horns are high intensity sonicators, but will the sample not heat up a lot during sonication? Heating of the sample during sonication, I heard, is supposed to denature the proteins, even break the formaldehyde bonds.
Thanx again both of you for your help!
Woe, this tip works and solved my foaming problem too! Previously I used a power setting of 15% (setting 3) and got foaming easily. After I lowered the setting to 10% (setting 2), I hardly got any foaming. Thank you Nick!
Here is my result with high and low power (volume 400 ul)
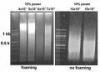
no problemo pcrman,
I always thought that you needed the high setting to ensure proper chromatin shearing. but you don't ![]()
N
I'm using sonication to lyse E. COLI in a protein purification procedure. I'm trying to explain some strange results I observed.
I used a 25ml sample in a sonication test on duty-60%, intensity=6 for 5 minutes, taking samples every minute. Running the crude extract and the insoluble suspension on an SDS-PAGE gel for each time point showed very little difference in sonicating one minute and sonicating five minutes.
In addition, the insoluble and soluble bands were identical, which makes the presence of inclusion bodies less likely.
My lab is relatively new to sonication, so the hope is that the error is ours and not in the experiment. If sonication produced foaming, therefore denaturing the proteins, would this explain the results - at least in part?
Hi mwwall,
Here we are talking about sonicating DNA. It seems that you are sonicating protein, right?
Once foaming occurs, sonicating efficiency goes almost to zero. I found your power setting is very high, have you experienced foaming? If yes, decrease your setting. Your are using 25 ml sample for sonication, that sounds to much to me. For ChIP sonication, the max volume I tried is 400 ul. Certainly the bigger the volume, the lower the efficiency.
