RT-PCR primer design guide - How to check gene structure and design the primer? (Nov/03/2003 )
Hello,
I would like to ask generally how you design your RT-PCR priemrs and specifically, how you check gene structure to contain some introns in the amplified region. Please let me know where is the best place to get such information.
thank you.
ZW
Hi ZW,
A potential problem with RT-PCR is DNA contamination in RNA. To tell whether amplication is from cDNA template or genomic DNA, RT-PCR primers are usually designed to span introns or bridge an exon-exon junction. When two primers span one or more introns (Fig.1), the amplified product from genomic DNA will be bigger than expected. Primers can also bridge an exon-exon junction (Fig. 2). Such designed primers will not amplify genomic DNA template because the intron will intervene the pairing between the primer and DNA. But, make sure that only a few nucleotides of the 3' portion should pair to the 3' exon as shown in Fig. 2. If the 3' portion of the primer has substantial pairing with the 3' exon, it can still initiate amplication without its 5' portion pairs to the 5' exon (Fig 3. sense primer). 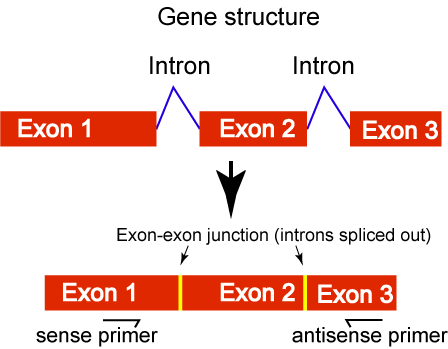
Figure 1. ![]()
Figure 2.![]()
Figure 3
This is how I usually design RT-PCR primers:
I. First get the cDNA sequence of my gene from NCBI. I only use Refseq if available. RefSeq represents the most reliable and unique cDNA sequence for a gene. Because searching the nucleotide database is not effective (you may get hundreds of sequences for a gene, most of them are irrelevant) so I will go to Entrez Gene database (previously LocusLink) at https://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene to find the gene of my interest, then scroll down to the NCBI Reference Sequences (RefSeq) section to find the RefSeq mRNA sequence whose accession number usually looks like NM_xxxxxxx. Then, click the sequence link which will lead to the actual sequence from GenBank Nucleotide sequence database.
2. Copy the sequence to a program to design primers such as Primer3 at https://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. Just accept all default parameters provided by the program and design primers. Choose the pair with highest score.
3. Copy the sequence corresponding to the amplified region by the primer pair you have chosen and then go to UCSC genome browser at https://genome.ucsc.edu/cgi-bin/hgBlat and paste the sequence there and do a BLAT.
4. Check if the amplicon spans any introns by view the detailed alignment of the query sequence against genomic sequence.
5. Or do a BLAT using the whole cDNA sequence first to know where exon-exon junctions (on cDNA) are and then design primers by specifying a target region corresponding to the junctions
6. Another purpose of Blatting amplicon sequence against genome database is to check if the amplified fragment has any homology with other genomic regions. The Blat results will tell that.
7. There are some other places you can also check gene structure such as NCBI Aceview at https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html and Ensembl at https://www.ensembl.org,
Edit: Sept. 6.
UCSC also provides an in silico PCR tool which allows you to input a pair of primers and retuns potential amplicon targets in the genome. It is useful for checking off-target amplification by your Rt-PCR primers. You can find the tool here: https://genome.ucsc.edu/cgi-bin/hgPcr
Hope that is of some help
Agree.
I have a question:
Which part of a cDNA sequence (5' UTR, 3' UTR, ORF, or any motif region) will you choose as amplifying target for RT-PCR, if they satisfy all the requirements (uniqe in genome, average GC, etc)? My feeling is they should all be OK for expression study. Am I right?
Kawaka
I have a question:
Which part of a cDNA sequence (5' UTR, 3' UTR, ORF, or any motif region) will you choose as amplifying target for RT-PCR, if they satisfy all the requirements (uniqe in genome, average GC, etc)? My feeling is they should all be OK for expression study. Am I right?
Kawaka
Is it right???
Hi All !
Notice that to study gene expression, the best is to design primers on two different exons, thus you obtain something specific for mRNA/cDNA, because you can distinguish cDNA from genomic DNA
Take a look here... this could help !
https://www.biocompare.com/techart.asp?id=799
Hi ZW,
first a warning: I'm from the company making this software.
But you could try www.probelibrary.com/adc which designs intron-spanning real time PCR assays on the fly. Only, you have to use Probelibrary Probes for these assays.
I would be interested in knowing your opinion of the tool...)
Søren
Søren M. Echwald, MSc., Ph.D.
-----------------------------
Exiqon A/S
Bygstubben 9
DK-2950 Vedbaek
Denmark
www.ProbeLibrary.com
One Real-time PCR kit, which covers 38.565 genes
Kawaka wrote:
I've had some problems with RT-PCR when using primers in the 5'UTR region. A gene may have different transcription initiation sites in different tissues, maybe even in a same cell line under different conditions. As an example, I was not able to amplify a gene from total pancreas RNA using the upper primer in the 5'UTR (nucleotides 69-80 of the RefSeq sequence) but the same primers worked wonderfully on liver RNA. I changed the primers to the exon2-exon3 region and then I got similar levels of amplification from liver and pancreas. It is not a case of alternate promoters, the exon 1 is the same in both tissues, it's only that different transcription start sites a few nucleotides upstream or downstream may be used preferentially in the two tissues.
Hope this helps. Cheers.
I have a question:
Which part of a cDNA sequence (5' UTR, 3' UTR, ORF, or any motif region) will you choose as amplifying target for RT-PCR, if they satisfy all the requirements (uniqe in genome, average GC, etc)? My feeling is they should all be OK for expression study. Am I right?
If you use oligo dT primer for RT reaction, it is advised to design RT-PCR primers half way to the 3' end of the mRNA sequence in case that the RT reaction for some genes doesn't extend long enough to the 5' portion.
It's everything very nice. I have one question though, probably simple. If my gene has pseudogenes, I mean with less or no introns, where do I look for their sequence, to design primers that do not amplify pseudogenes? However hard I can try to get rid of DNA, there's always a possibility that there's something left and the pseudogene will amplify instead, especially if it's a low-copy gene and I put a lot of cycles.
...
You can also try AutoPrime at www.autoprime.de.
It does the same thing but gives you the option to change the parameters, choose different output formats, etc.
Greetings,
Flexer.
