SDS-PAGE irregular and difuse bands - what's your opinion? (Jun/26/2009 )
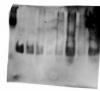
This is a western, where the part of the western appears to work.
However, the SDS-PAGE is weird. In all the experiments, in almost all cases (but not all cases) the band front is very irregular, very diffuse. However, in all cases the markers (coloured ones) elute perfectly (you can see sharp bands, even when its front has been horrible).
In the image the markers are not very visible (1st lane), but I'm worried about the protein bands
25 kDa protein
lane 1 - markers
lane 2 - 10 ng
lane 3 - 5 ng
lane 4 - 3 ng
lane 5 - 1 ng
lane 6 - 3ng in 100 µl H2O, dried with speed vac, resuspended with HEPES
lane 7 - 10ng in 100 µl H2O, dried with speed vac, resuspended with HEPES
lane 8 - 3ng in 100 µl H2O, dried with speed vac, resuspended with PBS
lane 9 - 10ng in 100 µl H2O, dried with speed vac, resuspended with PBS
So, I have these questions:
1) why the bands are so diffuse
2) why the protein appears to give an enormous trace in some cases?
3) why the front of the bands is horrible, although in many cases after I see sharp bands (not this case)?
I don't know what it can be. One person in lab used my same solutions (all of them) and he had a perfectly defined front band. I've had sometimes a perfect front band, but I don't control why.
Well, this is a 10% acrylamide gel made in lab, 160V.
Anybody has any idea from the picture?
i would look at the buffer(s) in which the samples are prepared.
what was the buffer in which you dried them?
what was the pH and concentration of the hepes? how did it affect the pH and ionic strength of the sample?
same with the pbs, with particular emphasis on the salt added to the sample?
in other words, your samples are more than likely the culprit.
But samples are simple standards, a recombinant protein at a given concentration, diluted in hepes (exactly, (10mM Hepes, 150mM NaCl, 3mM EDTA, 0.005% Tween-20), or diluted in PBS (Sodium chloride, 150 mM, and sodium phosphate, 150 mM).
Since working with the same samples, I've seen very sharp bands once stained (or by western) although it had a horrible front band during the electrophoresis, and I've seen the against, and the other two combinations as well, I think that this should not a problem of my samples.
I don't know, maybe the gels (done in-lab) are better polimerised some times and this explains everything? I'm sceptic with the voltage, I've run gels at 350 V and the bands were perfect. Now I work at lower voltages, 120 V or 160 V in function of the time I have, but I have to discard the voltage as a possible cause.
Perhaps is the running buffer? Bad idea to re-use it? But with fresh buffer I've had the same problem many times, so I should discard this as well.
I though it was the sample load buffer, low content of glicerol maybe, but other people has used without any problems (this points to my samples, yes, but I've had perfect bands with them some times!).
I don't know, are there a problem with the samples? Indeed, the recombinant protein has excipients and is resuspended in H2O. From that solution, I dilute 1 µg in 1 ml of the hepes buffer. In the band in the gel corresponding to 3 ng of protein dried with speed vac, there is 3 µl of the hepes solution in 100 µl of water, dried and resuspended in 5 µl of PBS (+ 5 µl Sample load buffer). So if we point to the tween20, I have 25 picoliters of tween20 in the solution. I can't believe that this is causing all of this.
But indeed, if the markers are always well-defined (although the front band is horrible), it is like the samples are the problem, isn't it?
I've though in applying the next modifications
- don't reuse running buffer
- use always fresh SDS (could this be the problem? some times I use SDS (10%) that has been 1 night at room temperature after defrozen)
- polimerise the gels with solutions WITHOUT SDS (I've read this improve ressolution)
- polimerise the gels the afternoon before the experiment (usually I use the gel immediately after the polimerisation, but I leave 40 minuts for both parts of the gel
- try to reduce the sample volume. Perhaps this is the cause of this horrible bands, I should work with smaller volumes
Let's see what I get.
mdfenko on Jun 26 2009, 07:21 AM said:
what was the buffer in which you dried them?
what was the pH and concentration of the hepes? how did it affect the pH and ionic strength of the sample?
same with the pbs, with particular emphasis on the salt added to the sample?
in other words, your samples are more than likely the culprit.
I'm running another gel. This time, the solutions for the polimerisation of the gel have not SDS and the double of salts concentration, with the aim of obtaining more ressolution. In addition, I've used 1.5 mm glasses to have less filled wells, although this time I've used the minimum volume possible. 120 V with fresh running buffer. However, although the front of the band is not irregular, this is quite diffuse. But I can see the markers perfectly.
Then, since I'm seeing the diffuse bands even with wells where I've only added sample load buffer. I hipothesise:
a_) I have a problem with the sample load buffer (but a partner used the same with their samples and the front was OK)
b_) I have a problem with the stacking.
What problem can I have with the stacking? I use the same solutions, the stacking at 4 % of acrylamide, the separation at 10 %.
you are resuspending your sample in pbs before adding sample buffer. this gives a relatively high salt concentration to the sample. this is not good for the sample. suspend in water before adding the sample buffer.
leaving the sds out of the gel should not make a difference if you have sds in the electrode and sample buffers (the old pharmacia phast gels did it this way so that they wouldn't have to make too many different gels, just different buffer strips).
check your stacking gel buffer and sample buffer pH. if they are not where they should be then you may have a stacking problem.
i still believe you have a salt problem.
I did migrate some samples with more than 500 mM of NaCl and never had any problem. Checking pH of your buffers is definitely a good idea though.
Please, tell me you solved it. Now I think i have the same problem, my resuts are like yours in the picture. I´ve checked a lot of things (products, samples, voltage, buffers) and it seems everything is ok, but not, there is something wrong in running or polymerization, but what?
I also believe i tmight be too high salt in the samples which can affect the whole gel, so also the wells with loading buffer only
had that problem with RIPA buffer total cell lysates...
try to desalt with Zeba colums before loading and see if that solves the problem:
Zeba Spin Desalting Columns and Devices, 7K MWCO
good luck :-)
Thank you for your advice, but i am completelly sure this is not the problem becouse i have run this samples before with good results. I´ve also run samples from other people and obteined something like the picture. Running buffer, SDS 10% and Trises are prepared by a technician and nobody has my results. It´s something i can not controll becouse sometimes gels are well and samples run well and sometimes not, and i think i do things in the same way. What is happening?
