PCR and gel purification problem - did I screw this up (May/10/2009 )
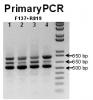
As shown in gel image (PrimaryPCR), this PCR primer produces 3 bands top and bottom one are the dominate bands.
The expect PCR products are the bottom one and the middle one (124 bp difference in size due to additional exon).
Now we don't understand what is the top one (around 850 bp) that is so strong?
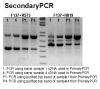
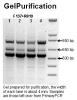
I purified the top one from gel (sample1 and 4 from first PCR). GelPurification is the image of gel I used for gel purification and use purified product as template run a secondary PCR. SecondaryPCR shows the gel image of second PCR results and although I only purified top band, I still got lower 2 bands using same primer (F173+R819).
My thought is that the relative large number of top band somehow prevents very few of lower 2 fragments from migrated to appropriate place and may likely trapped with top bands. That is why I still got traces of 2 lower bands at secondary PCR because they also got purified with top band.
I am very sure I DID NOT touch any of lower bands because I run another longer gel for gel purification and the distance between top and middle band is at least 3mm I donít think anyone can miss that (as shown in GelPurification).
My boss did not buy my idea.
During second PCR, I also try a difference primer pair (F137+R573) that the reverse primer move about 100 bp (near 250 bp of include 124 bp insert) upstream which will not detect additional 124 bp fragment. The result shows the bottom band shift downward about 150 bp and no middle band identified; however, the top band remains at same position even use gel purified product (F137+R819 PCR product).
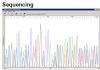
I also send purified PCR product from first PCR for direct sequencing and result indicates this purified product contain 2 equally strong templates (as shown in Sequencing). The sequence alignment and fragment search suggest that this band probably has nothing to do with our target gene (2 lower bands).
My boss simply thinks I screw up during gel purification; and the evidence are I got lower bands during secondary PCR and multi template of sequencing.
I try to explain myself (stupid thing to do to a big professor), and my boss end our argument by giving me 2 weeks notice. I am going to leave anyway, I actually glad it end sooner.
I think you have a half/half situation here: I would say that, yes, there probably is a contaminating band in your gel extraction, but that you probably have a PCR that mis-primes as well, so that you get the smaller bands in your gel after the second PCR.
A large PCR product is not enough to prevent smaller products from moving through a gel, even genomic DNA will not prevent this, unless there is a massive over-loading of the DNA onto the gel.
bob1 on May 10 2009, 05:40 PM said:
A large PCR product is not enough to prevent smaller products from moving through a gel, even genomic DNA will not prevent this, unless there is a massive over-loading of the DNA onto the gel.
Thanks for suggestion
We are doing alternative splicing so we always have multi-bands from single PCR. May be PCR God does not like me, I often have purified larger fragment still containing smaller fragment.
That is why I have thought that larger fragment prevents few smaller fragments from moving, since only hundred or few dozen smaller fragments would be sufficient to get visible product during second PCR.
Someone in this forum suggest this could be a product dimmer. Can product dimmer form with PCR products that have nothing to do with each other.
I have also try additional PCR to test the primer, and I found out that single forward primer (F137) along is enough to produce product that is about 850 bp. Guess it solve the question why I have 2 equal strong template in sequencing result.
wuxx0153 on May 11 2009, 08:13 AM said:
bob1 on May 10 2009, 05:40 PM said:
A large PCR product is not enough to prevent smaller products from moving through a gel, even genomic DNA will not prevent this, unless there is a massive over-loading of the DNA onto the gel.
Thanks for suggestion
We are doing alternative splicing so we always have multi-bands from single PCR. May be PCR God does not like me, I often have purified larger fragment still containing smaller fragment.
That is why I have thought that larger fragment prevents few smaller fragments from moving, since only hundred or few dozen smaller fragments would be sufficient to get visible product during second PCR.
Someone in this forum suggest this could be a product dimmer. Can product dimmer form with PCR products that have nothing to do with each other.
I have also try additional PCR to test the primer, and I found out that single forward primer (F137) along is enough to produce product that is about 850 bp. Guess it solve the question why I have 2 equal strong template in sequencing result.
You may try semi-nested or nested PCR on amplified DNA with one of the primers or both being specific to the expected product
Good Luck
wuxx0153 on May 11 2009, 05:13 AM said:
bob1 on May 10 2009, 05:40 PM said:
A large PCR product is not enough to prevent smaller products from moving through a gel, even genomic DNA will not prevent this, unless there is a massive over-loading of the DNA onto the gel.
Thanks for suggestion
We are doing alternative splicing so we always have multi-bands from single PCR. May be PCR God does not like me, I often have purified larger fragment still containing smaller fragment.
That is why I have thought that larger fragment prevents few smaller fragments from moving, since only hundred or few dozen smaller fragments would be sufficient to get visible product during second PCR.
Someone in this forum suggest this could be a product dimmer. Can product dimmer form with PCR products that have nothing to do with each other.
I have also try additional PCR to test the primer, and I found out that single forward primer (F137) along is enough to produce product that is about 850 bp. Guess it solve the question why I have 2 equal strong template in sequencing result.
Hey wuxx0153
How did you solve the problem?I have similar problem.While I was seeking for similar topics to talk about my problem, I saw your topic.
Could you tell me roughly if you found where the bands came from?My double digestion is OK. Before ligation, after gel purification,I run quantification gel.Addition to my vector, I see the insert that I didn't purify from gel, on the quantification gel.
