Serious issues with RT-PCR - (Mar/20/2009 )
Hi all,
First of all this is my first post here, so hello, and glad have found this great tool!
Ok, to the problem. For some time now I have been trying to do RT-PCR - isolate total RNA from cells, reverse transcribe it using random hexamers/nonamers, and PCR amplify transcript of interest. Unfortunately, to no avail... Perhaps you can help me make sense out of it:
1. The RNA purification using TRIzol reagent looks good. I spec my rna's and also look at them on denaturing gel. Concentrations are usuall 1-2ug/ul, and 260/280 hovers 1.8-1.9.
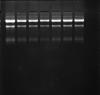
2. I tried 2 different methods for first strand synthesis on my RNA (2ug) - NEB First Strand and INVITROGEN SuperScript First strand. I'm not sure how to test for successful cDNA syntheisis, but:
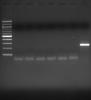
3. ... PCR for either my genes of interest or some housekeeping gene (B-actin is our usual) using 2ul (out of 20ul RT reaction) is never successful.
4 However, if I take cDNA (identical experimental samples) from another lab mate, prepared with the same reagents and same protocol, I do get PCR product - far right lane (so, PCR not an issue):
5. HOWEVER, if I take RNA from my lab mate (which looks identically on the denaturing gel and has similar spec values), I can do RT and PCR and get a successful product! (alternating hers or mine RNA's as a starting material):
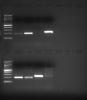
So... seems like there is "no problem" with either RNA purification, RT, or PCR... Except that if I use my RNA to start with, I cannot get any product!
So, assuming that RNA has good OD's, fairly high concentration, and looks good on denaturing gel, what else can be WRONG with it that prevents successful RT-PCR? Please, help me figure this out as I completely ran out of ideas by now... ![]()
maybe your rna preparation has something that inhibits reverse transcription?
what is your rna in?
I don't think so. After I wash the pellet with 75% Ethanol I let it air dry for 10 mins and resuspend in 60-80ul of DEPC-water (Fisher).
can you try poly-dT priming?
mdfenko on Mar 20 2009, 03:11 PM said:
I did once (with NEB reagents), but it didn't change the outcome. However, I can't use poly-dT for experiment since I'm looking for both poly-A and non-poly a RNA particles (both viral transcripts and replication templates of RNA virus), so only random primers will let me get cDNA for both.
maybe you still have some ethanol present?
can you let it dry longer?
mdfenko on Mar 20 2009, 03:35 PM said:
can you let it dry longer?
I can try... But usually after 10-15 minutes most of the pellets look pretty dry and are very hard to resuspend in water. They go from whiteish-wet to clear/glassy dry. Do you think that absolutely trace amounts of ethanol like that (pretty dry pellet into 80ul water; 2-3ul of that into 20ul reaction) would completely inhibit RT activity? It wouldn't be more than 0.1%-0.01% final ethanol concentration if the pellet wasn't completely dry, and TRIzol protocol asks not to completely dry the pellet.
walmark on Mar 20 2009, 12:27 PM said:
So... seems like there is "no problem" with either RNA purification, RT, or PCR... Except that if I use my RNA to start with, I cannot get any product!
So, assuming that RNA has good OD's, fairly high concentration, and looks good on denaturing gel, what else can be WRONG with it that prevents successful RT-PCR? Please, help me figure this out as I completely ran out of ideas by now...
Looking at the last gel, it seems that at some point between the RNA gel and the RT reaction, you are getting some RNA degradation that your lab mate is not. Go through with her everything you do different in that time, step by step. Are you using different stocks of water? Are you freeze/thawing any more times than her? Are you using a different freezer? Are you any less careful about RNAse (cleaning your bench, using RNA-designated filter tips etc)? Is there a vent above your bench blowing RNAse-laden dust down into your samples?
Also check whether you use different stocks of reagents for the RT rxn. In particular, look at dNTPs. The rest of the stuff that goes into a RT rxn is only used for that, so the tubes are only opened when you're being very careful to avoid RNAse contamination. Tubes of dNTPs, on the other hand, may have been used for PCRs or other procedures in which RNAse is not a concern.
It could also be, like mdfenko said, something in your prep. Trizol is classic and cheap, but personally I have never had a Trizol prep work as well in RT-PCR as a silica-based column prep. I think the biggest problem is getting all the salt and then ethanol out of the pellet. You have to be really good to do that well and not seriously sacrifice your yield.
So I'd say first check to see if your doing something that's degrading your RNA. If you don't find anything major, get some RNeasy (or equivalent) columns from someone on your hall and try getting your RNA that way. It may not be as satisfying as pin-pointing your problem, but I bet it will work a lot better for you.
