RNA isolation form HUVEC - UGLY RNA!! (Feb/14/2009 )
Hi people!!
I've been extracting RNA from my HUVEC cells, do they look ok? I'm familiar with fungal RNA, but the HUVEC RNA doesn't look quite like I expected. The 28S band doesn't seem like it is 2x intensity of the 18S band. I used the Qiagen RNeasy Minikit, used the buffer RLT + B-mercaptoethanol added to monolayer and pipetted several times. Then pipetted into microfuge tube and vortexed 30sec. Then continued with the rest of the kit protocol.
I've also tried Trizol, yield is higher but protocol takes more time so I prefer the Qiagen kit. Is there anything I've done wrong? Only I haven't done DNase digestion, plan to do that today and run another gel and see.
First picture's Trizol has nothing, I don't know what happened there. 2nd pix is another batch of HUVEC RNA + Candida RNA run on fornmaldehyde gel. 3rd pix is the same RNA run on normal agarose gel.
All RNA reverse transcribed and PCR with b-actin gave correct bands.
I use 5x104 cells per well.
Thanks!!
Chris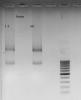
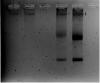
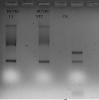
The RNA looks right in the normal gel. Maybe your formaldehyde had gone bad.
The RNA looks OK to me too. If the PCR is working is there really an issue?
bob1 on Feb 19 2009, 07:26 AM said:
If the formaldehyde had gone bad then the RNA for the Candida sp should not be so clear. The Candida RNA is very nice.
Yes purity and RNA integrity is an issue cause I'm going to use it for microarray.
I've had suggestions that my cells are not enough, and that I should extract from a T25/T75 flask instead of the 24-plate wells. But purity and integrity should be good no matter the concentration right?
Please any other ideas? maybe I should change my kit reagents to new ones. But then the Trizol method also gives lousy bands, even the 28S becomes blur.
Thanks
Chris
Try add some NaCl (10mM final would be good) to your loading buffer, then heat and load to the gel. Sometimes difference in ion binding to the RNA (and DNA) could make a big difference in its gel migration pattern. Your yeast RNA's salt content might be different from the HUVEC RNA.
