Trouble transforming pET28a in E. coli BL21(DE3) - (Feb/04/2020 )
We are facing issues while transforming pET28a vectors in expression strains. Please refer to the image attached and suggest improvements.
The quality of vector DNA and transformation efficiency in Top10 is good. All the suggestions will be appreciated
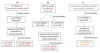
How many times have you observed this phenomenon and which plasmid extraction system are you using? If you are doing manual minipreps (i.e. not kit) it is possible that the DNA is not clean enough to get a good transformation and resulting in no plasmids coming out the final step.
bob1 on Wed Feb 5 02:10:39 2020 said:
How many times have you observed this phenomenon and which plasmid extraction system are you using? If you are doing manual minipreps (i.e. not kit) it is possible that the DNA is not clean enough to get a good transformation and resulting in no plasmids coming out the final step.
we have observed this particular phenomenon 3 times. The plasmid isolation method is manual and the plasmid quantity and quality is good (Verified by nanodrop and quantus). The same plasmid can be transformed in Top10 but not in expression strain. Other plasmids are also isolated by this same method and they are getting transformed properly. This problem is only seen in case of pET28a vector.
Based on your flowchart it seems you have done most of the appropriate experimental controls to eliminate bad plasmid preps and “incompetent” BL21(DE3) competent cells as the problem. When you say transformation failed, do you mean that you get no colonies at all? Or you get colonies but they have incorrect (deleted) plasmids?
I’m not an expert with this Novagen plasmid/host system. I’d revisit the Novagen manual and make sure you have the correct Vector/Expression host pairing. I took a quick look and BL21(DE3) is indeed the most commonly used strain, but there is one called BLR (DE3) that is a recA- derivative of BL21(DE3) that may stabilize some target genes with repeats. You also have to consider the possible toxicity of the expressed protein on the bacterial cells. Though that would not explain why you can’t get empty vector into the expression host. Also, when you get stuff from other labs (part A of your chart), you do have to consider the possibility something was not labeled correctly and what you got was not what you think it is. So I’d do some RE digests and make sure the plasmid you are using is what it is supposed to be.
I am baffled, as are you, at experiment B with the commercial construct- 1st generation transformation successful with both hosts, 2nd gen transformation not (but only for the DE3 strain). Did you try the TOP10 cells for that 2nd transformation? That would have been an important comparison.
I hope you figure this out and if you do, let us know!
OldCloner on Thu Feb 6 19:59:20 2020 said:
Based on your flowchart it seems you have done most of the appropriate experimental controls to eliminate bad plasmid preps and “incompetent” BL21(DE3) competent cells as the problem. When you say transformation failed, do you mean that you get no colonies at all? Or you get colonies but they have incorrect (deleted) plasmids?
I’m not an expert with this Novagen plasmid/host system. I’d revisit the Novagen manual and make sure you have the correct Vector/Expression host pairing. I took a quick look and BL21(DE3) is indeed the most commonly used strain, but there is one called BLR (DE3) that is a recA- derivative of BL21(DE3) that may stabilize some target genes with repeats. You also have to consider the possible toxicity of the expressed protein on the bacterial cells. Though that would not explain why you can’t get empty vector into the expression host. Also, when you get stuff from other labs (part A of your chart), you do have to consider the possibility something was not labeled correctly and what you got was not what you think it is. So I’d do some RE digests and make sure the plasmid you are using is what it is supposed to be.
I am baffled, as are you, at experiment B with the commercial construct- 1st generation transformation successful with both hosts, 2nd gen transformation not (but only for the DE3 strain). Did you try the TOP10 cells for that 2nd transformation? That would have been an important comparison.
I hope you figure this out and if you do, let us know!
Failed transformation means no colonies observed. Also, I tried TOP10 cells for the 2nd transformation and it was successful. I am also puzzled on how to proceed with this problem. I will keep posting on this thread regarding the progress.
This may have nothing to do with it, but what is your selective drug in the agar for transformation? And at what concentration? I had a weird situation once where a client had been trying to clone something in a high-copy plasmid that normally allows survival on Amp 100 mcg/ml. It should have been a simple subcloning, and they had tried over and over with no success. They asked me to take a shot at it. I typically use Amp at 50 mcg/ml in my agar, and the cloning worked perfectly on my 1st attempt. When I gave them the successfully transformed clone, they told me it was dead-they could not grow it! But I could subculture it with no problem. Finally we figured out that it would not grow on their plates because, with the insert, it could not tolerate the higher conc.of Amp, but would tolerate the lower conc. Why? We have no idea. It was just plain weird. So maybe you can experiment with lower conc. of your selective drug? Just a shot in the dark.
