Albumin isoforms - (Oct/03/2016 )
Hi I wondering if anyone out there could help with this.
I'm looking for autoantibodies in a healthy group of patients Vs disease group.
I've done some immunoprecipitation using IgG extracted from patient blood. I ran the IP out on a 2D gel and then had the spots identified by mass spectrometry. Our results indicate that there are a lot of albumin isoforms on the 2D gel and that they are reacting with the IgG.
Indicating that the patients are producing antibodies against these various isoforms of albumin.
I need to verify this to be sure. Most of the isoforms (based on the sequences identified by MS) seem to be missing their N-terminus.
In terms of buying in an anti-albumin antibody to verify my work - Would an anti-albumin antibody that has been raised against the entire sequence of albumin bind with cleaved isoforms of albumin? Or will it only ever bind to the full sequence?
Anyone have any other ideas how I go about verifying the MS results?
many thanks
The question is - where did the albumin come from? It's pretty common to use bovine serum albumin as a blocker in IP experiments, so it might not be significant that the IgG is binding albumin other than it diminishes any response that you can see as a true reaction. There is precedent for antibodies to non-specifically bind to proteins, including BSA, in IPs and other immuno-assays.
Hi, There was no BSA anywhere in this experiment.
IgG was extracted from patient plasma.
This IgG was then added to a protein lysate of proteins extracted from human synovium.
The resultant "captured" proteins (those that bound to IgG) were then separated on a 2D gel which was silver stained.
Spots were then cut from the gel and identified by mass spec.
There were numerous spots (all low molecular weight) identified as albumin.
a polyclonal anti-whole albumin should recognize several epitopes which may encompass all of the isoforms you found. you should determine this empirically.
Many thanks for your sound advice as always mdfenko
Mdfenko - I wonder if you would have any input on the following issue I am having trouble with.
I am trying to find (novel) autoantibodies. I am extracting IgG from patient plasma and adding it to patient synovial fluid proteins. In theory if there is any antigenic material in the synovial fluid they will bind to the IgG (we're not looking at IgE or IgM at present). I then pull out my ab/ag complex with A/G beads, boil in loading buffer, rehydrate in 2D lysis compatible buffer and run on a 2D gel.
I then have spots identified by mass spectrometry.
The problem is to date that we are only ever picking up IgG fragments and no other proteins at all. Just lots and lots of IgG frags.
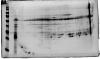
What am I missing here? Do I need to try and dissociate the antibody/antigen complex and remove the IgG so that I am only running the captured antigens on my gel? You can see from the attached image that there is a lot of IgG on the gel. The other alternative is that the IgG simply isn't binding to anything in the protein lysate (but I don't think that is the case)
how are you extracting the igg? it may be getting denatured during the process. the only "fragments" of igg you should see are intact heavy and light chains.
have you confirmed the binding with a non-denaturing gel?
have you run the complex on 1d?
have you run synovial fluid alone?
Hi mdfenko
The IgG is being extracted using Amicon® Pro Affinity Concentration Kit Protein A.
Would it not be standard to see lots of fragments on a 2D gel as the very nature of 2D PAGE will be to denature?
I have run it on a 1D gel. IgG only gives the expected bands at 150kDa (whole molecule), 50kDa (heavy chain) and 25kDa (light chain).
When I compare the 1D gel side by side to the immunoprecipitate there are extra bands - suggesting something has bound. However, as I said when I run the IP complexon a 2D gel all I'm getting are lots of IgG frags.
Maybe I need to try and separate the antigen off the IgG and run that on a gel to try and limit all the "background noise" I'm seeing from the IgG?
I haven't run the complex on a non-denaturing gel - what would be the advantage of doing that?
thanks!
the advantage of running on non-denaturing would be to determine if you indeed have a complex.
standard (or, at least, routine) 2d page is ief in the first dimension and sds-page in the second. ief is non-denaturing, separating based on pI (pH where net charge of the molecule is zero). sds-page is, indeed, denaturing but not fragmenting.
the complex should exhibit a pI different from the pIs of the separate proteins. then, on sds-page, you should see the breakdown of the complex to heavy and light chains of igg and the protein which had been bound.
you may see shifts in pI depending on number of proteins bound (if not 1:1) and/or degree of aggregation of the complexes.
Hi Mdfenko
Thank you once again. I plan to bind my IgG covalently to the beads using DMP. I am hoping then that when I do 1DE or 2D SDS PAGE I won't see any (hopefully!) IgG fragments as they have been retained on the beads and in theory I should just see the antigens.
My question about this is - My protein source that I will be using for the IP is synovial pellet proteins in 2D lysis buffer (urea/thiourea/CHAPS/DTT/ampholytes). If I add my IgG (which is covalently bound to the beads) to this protein lysate - are the lysis buffer components likely to "chop up" the IgG into fragments? This is what I want to avoid. I hope to only run the captured antigen on the gel.
Also can I just use the standard reducing buffer of SDS/tris-cl/DTT and boiling to elute the antigen? Or - would this buffer be too harsh and also elute the bound IgG also?
Sorry - lot's of questions!