phospho-creb western blotting - (Jun/15/2016 )
Hi
I seem to have serious trouble with p-creb blotting... I have tried numeorus protocols and using different Abs but I either get no signal at all or multiple bands which are pretty close and make it hard to really figure out what band actually represents p-creb. I would appreciate if someone could take a look at the attached film and help me figure out which one of the bands is my protein of interest. I've been using cell signal p-creb antibody which is suppose to be 43 kDa..ANY help or comment is welcomed since I am at a point where I'm pulling my hair out
THX!
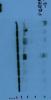

Its a little hard to figure out - it would help if you labeled the marker with size. Note that phospho-protein bands are often smears or stepped/multiple bands (depending on resolution) as additional phosphates get added. I don't know if this is the case for CREB though.
Your westerns look like they need some optimization - have you titrated the antibody to see what happens if you increase/decrease antibody concentration? details of what you have tried will help us to see what might be going wrong.
You may be able to get (phosphylated)peptide to block binding of the antibody, which would indicate which is the specific band.
bob1 on Wed Jun 15 21:49:09 2016 said:
thank you for your reply. I've attached the ladder I used for the membranes. As for your optimization suggestion - my lab is pretty tight on budget so I really ought to try and get the most possible out of these films. However, It might be helpful to understand the reasons for getting so many bands and whether this affects the validity of my results (assuming I am able to find the correct pcreb band). Also, Is it possible for a protein to show up higher in the blot than the specified molecular weight?Its a little hard to figure out - it would help if you labeled the marker with size. Note that phospho-protein bands are often smears or stepped/multiple bands (depending on resolution) as additional phosphates get added. I don't know if this is the case for CREB though.
Your westerns look like they need some optimization - have you titrated the antibody to see what happens if you increase/decrease antibody concentration? details of what you have tried will help us to see what might be going wrong.
You may be able to get (phosphylated)peptide to block binding of the antibody, which would indicate which is the specific band.

Yes, it is possible for proteins to run not at the predicted size - actually it is very common. I don't know if pCREB does this though. From a quick google image search it looks like it might run slightly higher than predicted, perhaps 45 kDa or a slight bit higher, so this might fit with the 48 kDa band you see. Forskolin treatment seems to enhance it, could you try that?
