RNA gel problem (melted bands) - (Jan/27/2015 )
Dear all,
I extracted RNA from Chlamydomonas cells with the traditional phenol:CHCl3 protocol (1X washing step) and precipitated it with LiCl (2.5 M final concentration).
Analysis at the Nanodrop gives good parameters and an amount of ~3 ug/uL (~3000 ng/uL), however when I loaded into the gel (5 uL + 1 stain), the gel after running (50 V, 50 min in TAE 0.5X) looks like the photo (MidoriGreen staining).
RNA degradation or some residual substances from extraction which interfere with running?
How is possible that the upper part of the run is without stain? Should I load less RNA?
Thanks!
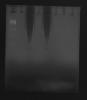
You will vastly overload the gel by loading 15 ug of RNA into a single lane. Dilute your sample 100x to 1000x and try again.
Thanks Phage! Yes, I overloaded the well, it's true..
But I had also a problem with TEA buffer because also DNA gels (right loaded) look completly wired.
