Difficult cloning/ligation/transformation blunt and sticky - (Dec/10/2014 )
Hi all, got a tricky cloning issue for you all to get your teeth into!
I'm trying to swap a small genetic fragment (400bp) in my vector for a longer one (2.2kb). It's a large parent vector (~14kB) and the enzyme sites are pretty unfavourable. The only way to excise the first fragment is with AscI and SmaI, which leaves sticky and blunt ends respectively. Thus I amplified my second fragment to have an AscI site at the 5' end, blunt at the 3'.
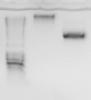
I digested the vector (see first image) and extracted the larger band. I then digested the successfully amplified insert and set up a ligation reaction.
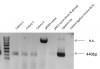
Post ligation I got several colonies on my plate. The first round of minipreps were digested with EcoRI along with the parent vector (image 2), but yielded very different digestion patterns when they should have been largely similar (i.e. with one band increased in size to account for larger insert). More colonies were tested, but with the same problem (~25 total).
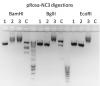
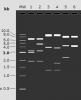
I repeated the ligation and transformation and got just 3 colonies. I subjected the colonies to colony PCR (see image), where the primers were targeted against the backbone and the insert and would be predicted to give a 440bp band. The 3 colonies tested all gave a band at the correct size, the parent undigested vector did not (but did give a larger non-specific band), whereas the vector that insert came from was also negative. I also tested the ligation reaction, which gave a strong band at the correct size, and the control ligation reaction which gave no band. So far, so good! (image 3)
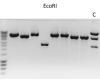
I miniprepped the colonies and digested with BamHI and BglII and EcoRI which all should give relatively similar digestion patterns to the parent vector (see predicted gel, image 4. Lane 1 control BamHI, 2 predicted BamHI, 3 and 4 BglII and 5 and 6 EcoRI). Again, all colonies failed to give predicted digestion pattern, but PCR confirms the correct insert orientation with the backbone. It appears there's a 'global' change in the vector structure. Has anyone ever seen anything like this before, or have any advice please? Everything seems to be working, apart from getting the correctly ligated vector into the bacteria!!
Thanks
A
(Also, I have just run out the backbone and insert I used for ligation to double check and they seem absolutely fine; see image, and have done colony PCR on the first batch where unsurprisingly the correct product appeared in all colonies (except the control colonies as expected))

Yep. I'm an idiot. SmaI has the same site as XmaI, but XmaI leaves a sticky end. New primer ordered!
