Missing DNA Target and Effect on Non-Specific Amplicons - (Jun/25/2014 )
Hello,
I'm working on mapping a large deletion in a canine model. I've used PCR to probe regions of WT-canine and Mutated-canine genomes to investigate where the deletion may begin/end. Seeing amplification in WT & Mutant indicates normalcy (outside deletion area), whereas amplification in WT and failure in the Mutant indicates probing within the deleted area. However, I've found an interesting result involving, what I believe to be, non-specific amplicons. Attached you see a gel image with identical PCR reactions, except the duplicates on the left contain WT DNA, whereas the duplicates on the right contain Mutant DNA. Ladder beings at 100bp, with 100bp increments. The expected amplicon size is 566bp, which fits what is observed in the WT lanes. So great, this area is most likely within the deleted area in the Mutant. However, what are these non-specific amplicons, and why are they much more prominent in the Mutant lane?
PrimerBLAST results in only one non-specific amplicon, with a size of ~1700bp (not present). Oddly, when I decrease primer stringency parameters, which should increase the reported number of non-specific amplicons, the one non-specific amplicon previously reported under default conditions vanishes. This makes me question the programs ability to detect non-specific amplicons.
However, in theory, if you were to design primers which had the ability to non-specifically amplify (reasonable number of mismatches), and then you were to remove its target binding region (such would be the case in my Mutant model), would you expect it to amplify the non-specific amplicons to a greater extent? I think this theory only fits IF the concentration of primer is limiting in the reaction equation, which may not be the case at earlier cycle numbers, but may be at higher cycle numbers.
For instance, say you start with 1 copy of your target, but you also have 5 non-specific targets. The target has an efficiency of 80%, while the non-specific regions only have an efficiency of 20%.
copies of target = 0.8 x 2n
copies of non-specific amplicon A = 0.2 x 2n
B = 0.2 x 2n
C = 0.2 x 2n
D = 0.2 x 2n
E = 0.2 x 2n
You can imagine, that by the time primer becomes limiting at later cycles, the intended target has much more copies than non-specific amplicons. This would "hog" primer. Not only that, but each non-specific amplicon produces a separate band - you have to get past a certain threshold to detect it on a gel.
What are everyone's thoughts? It's possible that something else is going on here (i.e. I'm within an inserted area in the mutant, etc..)
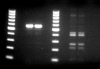
To test me theory, I ran a PCR at 2x the normal primer concentration. We'll see what the results show.
