Trouble with PCR of short sequence - (Jun/10/2014 )
Hi all,
I'm having problems with PCR for a few short sequences. Basically I want to identify what isoforms of the protein of interest are being expressed in the cell.
So what I've done is,
I used NCBI to design the primers by inputting the sequence I want amplified up and have double checked that it binds to the sequence.
I am using cDNA that has been converted from RNA extraction.
I have checked the Tm of the primers and they are about 50C, so according to New England Biolabs I reduced the annealing temperature from 55-->45C.
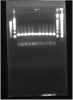
I've never really had any problems with PCR before, but this is my first time amplifying up such short sequences (50-100bp) so I'm not sure whether there are tips or tricks for dealing with short sequences. From the gel (1.5% agarose) I can see the GAPDH control band clearly at around 100+bp. So clearly the PCR method is working, but why can't I see the bands that I want?
Any advice would be appreciated.
PPAW
try a gradient (starting at 40C or lower) for annealing. 45C may be too high under the conditions of your pcr.
PPaw,
Are you saying that your PCR works fine with the positive control but you are not seeing any bands for your actual samples?
If yes, then you must look into you cDNA preparation and confirm that it is going well.
Could you label the gel and the marker used? Are you multiplexing your PCR by including GAPDH and your gene-of-interest primers in the same reaction?