If I am running Real-Time PCR with one gene of interest and one reference gene(G - (Apr/30/2014 )
I am testing many gene of interest, and I choose Gapdh as my reference gene. For some genes, I get one nice single peak, but for some genes, I get two peaks for my melt curve. Can I know why? Is it normal?
Maybe there is allele variability for that particular amplicon region. You could run the samples on a gel and see what the size difference is. If you see two distinct bands, you probably have allelic variation, if you see a smear adjust your cycling conditions. Are these previously established qPCR primers? Taqman?
jerryshelly1: He's talking about melting curve, so he must be using SYBR Green.
First of all it's not normal and it not good. SYBR assay must have only single peak, otherwise the global flurescence emitted by intercalator dye is combination of both products, which you can't quantify.
Thera can be many reasons for multiple peaks, first it's dimers (in that case one of the peaks has much lower melting temperature and moust likely is present in non-template control too).
Second reason is if primers span a short intron, the second product can be contaminating gDNA amplified. Some samples may have this peak, DNase treated samples would no.In this case you can try amplifying DNA, if it does make a peak of similar Tm.
Third possible reason is non specific aplification. That would be nonspecific accross all samples, but may give various different nonspecific peaks.
There may not be always visible more bands in cases of multiple melting peaks, since they may of very similar length just varied in the GC content enough to display different peaks (similar, rarely single melting peak may actually hide two products of different size). SYBR product are usually around 200, but Taqman assays may be too short to be even suitable for agarose gels.
Changing cycling conditions may help to get rid of non specific amplification, possibly even dimers in some cases, but not the amplifcation of DNA. Then you either need to redesign primers (which you should in any pother cases, where you can't get single product by any cycling modifications) or treat RNA with DNase (or with better DNase if you already do).
Trof on Wed Apr 30 22:01:32 2014 said:
jerryshelly1: He's talking about melting curve, so he must be using SYBR Green.
First of all it's not normal and it not good. SYBR assay must have only single peak, otherwise the global flurescence emitted by intercalator dye is combination of both products, which you can't quantify.
Thera can be many reasons for multiple peaks, first it's dimers (in that case one of the peaks has much lower melting temperature and moust likely is present in non-template control too).
Second reason is if primers span a short intron, the second product can be contaminating gDNA amplified. Some samples may have this peak, DNase treated samples would no.In this case you can try amplifying DNA, if it does make a peak of similar Tm.
Third possible reason is non specific aplification. That would be nonspecific accross all samples, but may give various different nonspecific peaks.
There may not be always visible more bands in cases of multiple melting peaks, since they may of very similar length just varied in the GC content enough to display different peaks (similar, rarely single melting peak may actually hide two products of different size). SYBR product are usually around 200, but Taqman assays may be too short to be even suitable for agarose gels.
Changing cycling conditions may help to get rid of non specific amplification, possibly even dimers in some cases, but not the amplifcation of DNA. Then you either need to redesign primers (which you should in any pother cases, where you can't get single product by any cycling modifications) or treat RNA with DNase (or with better DNase if you already do
Will we get one peak for GOI and another peak for Gapdh? Because for some of my experiment, Gapdh one peak, GOI is another peak.
Do you know how melting works?
SYBR Green binds to all double stranded DNA, therefore is non-specific. To better distinguish if you have a single or multiple product you run a melting curve. During a slow temperature increase in time all PCR products slowly dissociate from double strand to single strand. This causes the loss of fluorescent signal, because SYBR doesn not bind single stranded DNA.
The original signal decrease is transformed into a derivative graph, where you see "peaks" in places the melting curve has it's negative derivative maximum.
As in this picture: Up is the real melting curve, down is a derivative graph showing two distinct peaks.
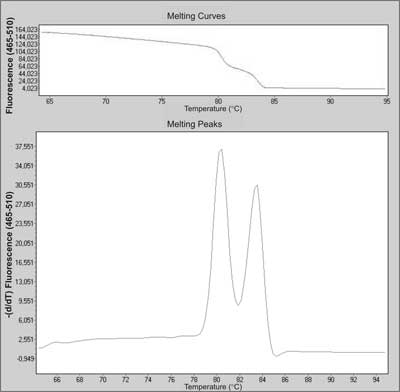
If you have a single product, it has homogenous melting profile and melting temperature depending on length and base content.
DIfferent amplicons have different Tm (defined as a top of the peak in the derivative graph), but have only single peak.
When there are more products, some of them "melt" in different temperatures, which makes a shift in the curve (as seen above) and then two local maximus = two different peaks.
So different genes can have different Tm , but should only have a single peak.
