Dna isolation from fungi - (Mar/31/2014 )
Hello,
I am trying to extract Dna from fungus (Alternaria alternata). I need really high quality dna for southern blots. Till now, i have tried a variety of methods, from CTAB method to qiagen kits. But, all i get is degraded dna or very low concentrations that are not usable at all. I always get a nice pellet but when i run the dna on a gel, it shows up as a smear.
Please help.
Thanks.
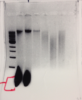
you have a picture of your gels?
Its hard to say how bad/good your results are to be honest and: smears are in general not that that weird to have when doing a genomic DNA extraction.
Here's a gel picture. The first two samples showed a huge Rna cloud, so i made sure to do a Rnase treatment after phase separation with chloroform. But then, the three samples showed only a smear. There's barely any dna in there even though my nanodrop showed a concentration of around ~500ng/ul.
Thanks.