vertical thin line from wells - ChIP testing of sonication efficiency - (Feb/22/2014 )
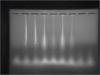 Hi, I am running a 2% agarose gel electrophoresis to test for sonication efficiency (chIP assay).
Hi, I am running a 2% agarose gel electrophoresis to test for sonication efficiency (chIP assay).
My problem is more of the gel electrophoresis itself - Why is there always a vertical thin line stretching down from the wells like that? Could it be contamination of some sort?
However, my ladder is newly bought and I'm quite certain it is not contaminated. The loading dye (6x) is new too.
For the first lane, I used 2ul Ladder + 1 ul loading dye, while the second lane i used 2ul ladder + 2ul dye.
For the rest of the DNA samples, I added 4ul dye + 20ul DNA, and loaded 20ul.
Any advice is greatly appreciated! Thanks!
Charlene
This looks as if you are making the agarose gel with water rather than buffer.
I made the gel with 1X TBE Buffer. Ran it with the same buffer as well.
do you dilute your ladder samples so that the final dye concentration is 1x?
in what buffer are your samples prior to addition of the dye?
what you are seeing looks like a salt effect.
I did not do so for my ladder, simply added 2ul Ladder + 1 ul dye. I'm not sure why the ladder didn't appear well either (first two lanes).
My samples are in ChIP elution buffer from the upstate millipore kit. I suppose it is made up of 1% SDS and 0.1M NaHCO3 as all homemade elution buffer recipes are.
I added 45ul of ChIP elution buffer to 5ul of sonicated chromatin. Then I heated the samples for 30 min with RNase A, and 2h with Proteinase K. Is that sufficient to get rid of the RNA and protein contamination? Could these be due to RNA and protein contamination?
What is a salt effect?
Thanks!
the "salt effect" to which i referred is the effect of relatively high ionic strength media on electrophoresis.
you may also be seeing an effect caused by pH variation in the sample, usually caused by the buffer in which the sample is residing overwhelming the buffer of the loading dye.
your ladder lanes are in a final concentration of loading dye of 2x and 3x, respectively. you should dilute to 1x loading dye.
as for rnase and proteinase k treatment, if you used the proper amount of each then the incubation times should be sufficient. but, heated to what temperature? how do you stop rnase? it may still be active during proteinase k treatment. how do you stop proteinase k?
mdfenko on Tue Feb 25 12:36:45 2014 said:
the "salt effect" to which i referred is the effect of relatively high ionic strength media on electrophoresis.
you may also be seeing an effect caused by pH variation in the sample, usually caused by the buffer in which the sample is residing overwhelming the buffer of the loading dye.
your ladder lanes are in a final concentration of loading dye of 2x and 3x, respectively. you should dilute to 1x loading dye.
as for rnase and proteinase k treatment, if you used the proper amount of each then the incubation times should be sufficient. but, heated to what temperature? how do you stop rnase? it may still be active during proteinase k treatment. how do you stop proteinase k?
I heated with RNase A 30 min, Proteinase K 2hours. I do not stop the RNase, I just add the proteinase K in after 30 mins. I dont know how to stop proteinase K either.. is there something I need to do? I usually just load immediately after incubation with proteinase K for 2 hours.
Okay, I've adjusted the ladder concentration and it turned out well! =)
Somehow, I re-ran the same samples and the lines didn't appear anymore! (lanes to the left of the middle ladder).
On the other hand, I heated some more DNA with 2ul of 5M NaCl for 4 hours, followed by RNase A 37deg, 30min, and proteinase K, 62deg, 2hours. The lanes on the right show the results for those samples. They actually look less promising than the left lane, which is really weird.
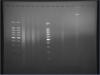
the ionic strength of your samples is too high. you may want to try precipitating the dna (after rnase and proteinase k treatment), washing out residual salt and resuspending in te or t buffer or water (if you use it immediately).
