Appearance of a higher molecular weight band after plasmid isolation? (NOT chros - (Jan/02/2014 )
Hello everyone,
I have a problem that I have never encountered before... I am cloning a couple of shRNA designed by the TRC guidelines to pLKO.1 vector. I constantly encountered this problem - I have a higher molecular weight band with the plasmid preps that I have done. I have tried Roche High Pure kit, isopropanol precipitation, phenol/chloroform extraction and PEG-precipitation as techniques and DH5a and Stbl3 as different strains. Previous colony PCR gave positive results but I could not get plasmid. Recently, I used the Macherey Nagel Nucleospin kit and obtained high-molecular weight band free plasmid...
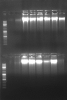
1 st lane is Fermentas GeneRuler Mix ladder, others are the shRNA directly after mini. The band is higher than expected but I have observed the same band in the Thermo Scientific "The RNAi Consortium (TRC) Lentiviral shRNA Technical Manual" (Figure 4), so it was allright. I put those into fridge.
Two days after (New Years' Eve and such), I tried to do a restriction digest with those. The gel is below. Upper row in the first gel is the left of the second ladder in the second gel, and lower row in the first gel corresponds to the right of the second ladder in the second gel.
Here, the plasmid band disappeared, and a higher molecular weight band appeared, which cannot be cleaved by the restriction enzyme pair that I previously used for screening pLKO.1 inserts (SpeI/NdeI). As controls, I had the "empty" vector that I have used for cloning (without the stuffer but still produces compatible ends), which is purified using Roche Genopure Midi Kit previously and there were no problems...
I am really perplexed by this, in my six years doing PhD I did not encountered any problems in cloning and now in my first post-doc I feel like I hit a brick wall. If you can provide me with any explanation on what may be going wrong I will be really glad.
Thanks,
Cihan Aydin
I realize this is an old question, and I hope Cihan figured out the problem. I had similar issues with both lentiviral and MoMuLV-based plasmids from my transformed colonies not cutting with enzymes on either side of where the insert would be, so I could not even tell how big they really were. So finally I cut with an enzyme that HAD to be there, the SspI that was in the amp-resistance cassette (Since the colonies grew on Amp, it had to be there, right?). This digest showed the construct to be way smaller than expected; some sequencing revealed that pretty much all the viral sequences had disappeared. Including the restriction sites that should have been around the insert. This kind of recombination can happen with viral integration vectors. Yes, even when the host cells are supposed to suppress such things. So try cutting with an enzyme in the vector backbone (rep origin, bacterial selection gene) that HAS to be present, and see what you get.
In my case I came to the conclusion that the insert was detrimental to the bacterium. I did finally get the construct, but it was only present in tiny microcolonies on the plate which I only saw after the plates had been stored a while. Any normal-sized colonies had deleted plasmids. Weird, huh?
There are some bacterial strains that can be used for unstable DNA, like Stbl2 to 4 from Invitrogen or SURE from Agilent or others. They are recombinase and/or endonuclease deficient or with other modifications. One of them is specifically designed for retroviral plasmids, but I don't remember which one. If the product is toxic, special vectors can be used that have no prokaryotic expression (even mammalian enhancers like CMV do express in E.coli to some extent).
It is all managable, but if you do not want to loose time and nerves, you may need a commercial solution. I grown a gene that was rummored to be complitated and recombined in E. coli in Stbl2 cells with (even Origene sent us a plasmid that was recombined, after we failed to obtain a full plasmid from cooperating lab, theirs was also deleted). Later also retroviral vectors. With no problems.