GAPDH Ct value - (Aug/06/2013 )
hello all
Recently i did qpcr and in this i found the CT value of GAPDH reaching 26.Its the first time i got a value like this. Most of the time i get at 18 or 19 Ct but now its around 26.
Ct value of the sample is at 28.
Do you think that there is some thing wrong?Could anyone please tell me why it is like this. If so does it affect while doing ablsoute quantification.
find the attached picture
thanks in advance
Hari
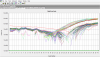
Can you upload the amplification plot with a linear y-axis instead of log ?
How did your other samples look like ? Can you compare with previous runs (has the Ct value of the sample become higher too, or is this just the case with GAPDH ?). Is it the same cDNA as previously ? If you have used the same cDNA as in previous runs and only GAPDH suddenly has a higher Ct, it does look like something is wrong.
Also, is GAPDH a suitable/established housekeeping gene for the cells the cDNA comes from ? Have you tried any others ?
Those curves look a bit funky.
It looks like you have a minor instrument issue too with that spike. If the cDNA is not normalized, then it doesn't matter if Gapdh is the same Ct value AS LONG AS you validated it prior using the same RNA input. Other options include using a reference gene panel to find a suitable reference gene. There is a good panel here. www.bio-rad.com/primepcr
As members before me mentioned that this graph are a little funky. There is starting to show a double waterfall effect so maybe the baseline is faulty (you need to correct it). And of course is GAPDH the best reference gene for your samples.
The second thing is that i once got double belly like graphs (on housekeeping gene) and the reason was that my temperatures of the run were wrong for 1,5°C.
regards
