What happen to my animal DNA extraction? - (Mar/21/2013 )
https://www.facebook.com/photo.php?fbid=566409006712238&set=o.224890614234699&type=1&theater
can anyone tell me what happen to my AGE result?
Actually this my laboratory practical session, so all lane is from same fish muscle tissues, just the protocols performed by different groups.
Can someone told me what happen for each land?..
which one is smearing and contaminant with polysaccharides, RNA or excess salt?
Thank you.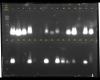
moved this thread here, as it's homework-related.
it would help if you told us what you are extracting, protocol, etc.
looks like you just have different loadings of small pieces of some nucleic acid.
you also need a ladder for the second row of samples.
ok.
fish muscle tissue as a starting material.
tissues washed with PBS buffer, then suspend in lysis buffer. (Tris, EDTA, SDS).
cold proteinase K is added and incubate in 55C waterbath for an hour.
phenol: chloroform is added--> centrifuge --> extract supernatant into new tube.
Then, 3M sodium acetate and absolute ethanol is added --> freezes for 10-15minutes at -20C.
then centrifuge again and dischard the supernatant, wash with ethanol.
the pellet then resuspend in ultrapure water.
finally load into 0.8% agarose gel.
mdfenko on Fri Mar 22 12:53:13 2013 said:
it would help if you told us what you are extracting, protocol, etc.
looks like you just have different loadings of small pieces of some nucleic acid.
you also need a ladder for the second row of samples.
may i know...are all the DNA band is smearing?
actually this is the first time for me to do the analysis, and i dont know why I get small faint band (lane 27)
is DNA contaminated?..
can you answer for me?..I am still learning how to in read the gel.
Thank you.
to me, the gel looks like you have an overloaded small dna band, not a smear.
how long is the dna sitting in ultrapure water before loading onto the gel?
ultrapure water (as with any water) will absorb co2 from the air and acidify. the acidified water will then cause dna to degrade. that's why we put our dna into tris (10mM) or tris-edta (10mM t, 1mM e), pH 8.
i assume that the idea of the exercise is to extract genomic dna. that makes the small size of the band somewhat distressing.
maybe i'm wrong. you may be extracting mrna from cytosol.
mdfenko on Fri Mar 22 15:51:23 2013 said:
to me, the gel looks like you have an overloaded small dna band, not a smear.
how long is the dna sitting in ultrapure water before loading onto the gel?
ultrapure water (as with any water) will absorb co2 from the air and acidify. the acidified water will then cause dna to degrade. that's why we put our dna into tris (10mM) or tris-edta (10mM t, 1mM e), pH 8.
i assume that the idea of the exercise is to extract genomic dna. that makes the small size of the band somewhat distressing.
maybe i'm wrong. you may be extracting mrna from cytosol.
I am not not quite sure how long dna sitting in ultrapure water, since after we (me & my fellow) resuspend in ultrapure water, we store it in fridge at 4C.
the gel loading is done by ours demo, since after the practical session, we still got another tutorial class (I am undergraduate student.)
For you information, this is first time we use ultrapure water to keep the dna, last time bacterial and plant dna extraction we resuspend it in TE buffer.
mdfenko on Fri Mar 22 15:51:23 2013 said:
to me, the gel looks like you have an overloaded small dna band, not a smear.
how long is the dna sitting in ultrapure water before loading onto the gel?
ultrapure water (as with any water) will absorb co2 from the air and acidify. the acidified water will then cause dna to degrade. that's why we put our dna into tris (10mM) or tris-edta (10mM t, 1mM e), pH 8.
i assume that the idea of the exercise is to extract genomic dna. that makes the small size of the band somewhat distressing.
maybe i'm wrong. you may be extracting mrna from cytosol.
what mean for overloaded small dna band? Not all dna side are same?
you are right, this session only to extract genomic dna.
And we need to discuss the result we get.
Really? lane 27 is extracting mrna from cytosol?..
I thought is DNA degraded, since there is almost faint at one side.
ASUKASIN on Fri Mar 22 16:08:10 2013 said:
I am not not quite sure how long dna sitting in ultrapure water, since after we (me & my fellow) resuspend in ultrapure water, we store it in fridge at 4C.
the gel loading is done by ours demo, since after the practical session, we still got another tutorial class (I am undergraduate student.)
For you information, this is first time we use ultrapure water to keep the dna, last time bacterial and plant dna extraction we resuspend it in TE buffer.
was the ultrapure water prepared fresh? if not then the dna may have degraded due to acidified water.
how many days was the dna stored?
ASUKASIN on Fri Mar 22 16:13:41 2013 said:
what mean for overloaded small dna band? Not all dna side are same?
you are right, this session only to extract genomic dna.
And we need to discuss the result we get.
Really? lane 27 is extracting mrna from cytosol?..
I thought is DNA degraded, since there is almost faint at one side.
genomic dna is very large and would migrate very slowly through the gel (it would remain near the origin).
fragmented dna, cdna, pcr products and rnas would move through more quickly.
due to your extraction technique, i wasn't sure if you would have broken open the cell nucleus to free the genomic dna (i think sds will but not sure). so i thought you may be looking at rna. mrnas are smaller than genomic dna and are seen as a smear. i think yours are too compact to be mrna.
however, i think you have very fragmented dna and may also be seeing some effect of salt on the migration (hence the appearance of the band).
mdfenko on Fri Mar 22 18:04:21 2013 said:
ASUKASIN on Fri Mar 22 16:08:10 2013 said:
I am not not quite sure how long dna sitting in ultrapure water, since after we (me & my fellow) resuspend in ultrapure water, we store it in fridge at 4C.
the gel loading is done by ours demo, since after the practical session, we still got another tutorial class (I am undergraduate student.)
For you information, this is first time we use ultrapure water to keep the dna, last time bacterial and plant dna extraction we resuspend it in TE buffer.
was the ultrapure water prepared fresh? if not then the dna may have degraded due to acidified water.
how many days was the dna stored?
ASUKASIN on Fri Mar 22 16:13:41 2013 said:
what mean for overloaded small dna band? Not all dna side are same?
you are right, this session only to extract genomic dna.
And we need to discuss the result we get.
Really? lane 27 is extracting mrna from cytosol?..
I thought is DNA degraded, since there is almost faint at one side.
genomic dna is very large and would migrate very slowly through the gel (it would remain near the origin).
fragmented dna, cdna, pcr products and rnas would move through more quickly.
due to your extraction technique, i wasn't sure if you would have broken open the cell nucleus to free the genomic dna (i think sds will but not sure). so i thought you may be looking at rna. mrnas are smaller than genomic dna and are seen as a smear. i think yours are too compact to be mrna.
however, i think you have very fragmented dna and may also be seeing some effect of salt on the migration (hence the appearance of the band).
I not sure when the ultrapure water is prepared but it keep in falcon tube in ours own lab table there.
I think DNA we only stored and few hours in 4C.
Based on the protocols, we used the LYSIS buffer to broke down the plasma membrane and nuclear membrane, and EDTA used to inactivate the nucleases.
if I have very fragmented dna, then mean for others group(lane) also have same with me? since all DNA at bottom part but not at origin.
If most are the fragmented dna, then what can I do to solve this problem?(in order to get high quality dna.)
