Genotyping PCR primers - (Apr/23/2012 )
I got a mouse from another lab with their genotyping primers, no info on which is forward or reverse and what I am sequencing. I got two products, as the sender of the mouse told me, one at 1300bp, one at 900bp. I have tried to align the primers to the gene and no luck.
So, I am running an experiment excluding one primer at the time, and further sending the bands for sequencing to see what am I seeing on gel. When I run the gel, I see the individual products but my positive control (which I know should have both bands from other gels) is not giving me the nice two bands, actually the upper band is just not there. To add to that, my lab moved just after I got the PCR working. In the new institution we switched from EthBr to GelRed, can this be why my positive control is not being able to resolve the two bands? So then, why the individual bands are resolved nicely? Thanks in advance for your help. I have done it three times with the same results or lack thereof.
Gel from left to right: Marker, Neg control, Positive control (containing primers 1, 2, and 3), Sample (with primers 1,3), sample (with primers (2,3), sample (with primers 2,1)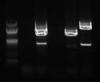
So you have the sequence for the primers, right, because you said
