PCR primer pairs - university exam - (Apr/05/2012 )
Hello,
just need some help with this question: I can't seem to make sense of iii> and iv>. To be honest, i don't think i understand the entirety of PCR in this context and why primers are even used?? but parts i> and ii> are pretty easy to work out from basic principle (i think - i got i> = Xa and Xf, and ii> = Primer C)
I know part iv is something to do with introns and exons, i think. lol! sorry. anyway here is the question -
Six mRNA transcripts (XA, XB, XC, XD XE, XF) that encode protein X isoforms are expressed in skeletal muscle from a single gene whose exon structure was determined. The figure below shows an alignment of the six cDNA (mRNA) sequences with exon boundaries indicated (vertical broken) lines are guides to show the alignment of exon boundaries). The arrows above XA in the diagram indicate four pairs of PCR primers, the arrows labelled with the same letter are a primer pair.
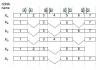
I basically need a simplistic explanation of what this diagram is in relation to PCR and like why primers are used. i'm pretty much a novice
Thank youu
Primers are used to recognise the target DNA. if one of the primers can't bind, there will be no amplification.
You are correct about part IV - but why???
genomic DNA has both introns and exons, cDNA only has exons. i'm guessing the target DNA is in the exons so it will be amplified in both genomic DNA and cDNA?
Yep, however, the introns in genomic DNA are usually quite long - too long for a standard PCR to work efficiently, so the other primer pairs won't amplify the genomic DNA, whereas pair C will.
There is something I do not get.
Why do you answer i> = Xa and Xf
Why not also Xc and Xe ?
Primer pair 1 can bind Xe and primer pair D can bind Xc.
Or am I missing something here?
Question is if it means only transcripts that are detected by both A AND D, or if the meaning is transcripts that are detected by A OR D. It's unclear.
i think it means together
does anyone know the answer or the explanation to iii) ?
iii) It's not that difficult to find out why. Look at the primers. Check if each primer pair even can amplify XB and if it does, does it detect only this transcript or any other?
Spoiler (select to display):
*
