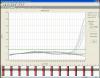
Strange RT-PCR amplification plots - (Jan/26/2012 )
Hi everyone,
I'm relatively new, but so far I have found this site very useful! However, I have a problem I couldn't find the answer to on the forums!
I have been getting some strange amplification plots for my RT-PCR recently. I use Taqman mastermix and tailor-made probes, which have worked in the past. But, having tried a new probe I have been getting strange results (see attached) - a hump-shaped curve initially, then a gap, then the reaction proper starts later on, but for most samples it doesn't reach a plateau before the cycles end. I add a "gene of interest" probe and a "housekeeping" probe into the same reaction, and the housekeeping one works fine, it's just the gene of interest that is a problem. I am using the same cDNA and the same reaction volumes as I have successfully used before.
Any suggestions? I have already tried increasing the cycle number from 40 to 50, adding twice as much cDNA and adding 10-fold less cDNA. Should I increase the cycles further, add more probe, or add more cDNA?
Thanks!
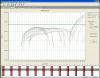
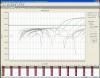
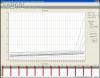
This is a logarithmic graph, can you post linear? Ideally a graph without baseline-correction.
I'll try ![]()
Those up the threshold look normal, but very late.
For those under there don't seem to be any start, they apper just flat, but this one is baseline-corrected curve, that could cover it.
There should be the raw output visible somewhere, it shouldn't have "delta" Rn in a graph. Maybe we will be able to see more. Unfortunately I don't remember well ABI interface but I know it's there somewhere.
If you have new probe, look specificaly for the initial fluorescence of those samples in raw graph, if it's higher that the previous probe it could be bad. Can you try SYBR assay to check the efficiency of the primers? To point out to the problem in the probe. We once get a bad working probe from ABI, they gave us new after we proved it.
If I saw those results, I would assume I had no template in the reaction. There is some amplification after 34 cycles, but that is often garbage rather than a real signal. Do you have a positive template control? Do you have evidence that your primers are working? There is very little to be gained in running more than about 35 -38 cycles.
Thanks for the input guys!
I'll take a look at the raw data when I can get on to the machine (the only computer with the software on!). Unfortunately my supervisor can't get more probe as he's short on money. But I'll see if I can borrow some SYBR.
As for positive controls, the cDNA I have used for the standards is supposed to express it highly. I ran a gel on the products yesterday and the bands were very faint compared to the ladder, but there looked like there were two products, maybe primer dimers?
Ok I've got some raw data.........
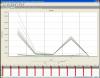
Hi everyone,
So I've tried doing the reactions in singleplex (i.e. without the housekeeping primer) at three different primer dilutions and there's been no change. The other options are increasing the annealing temperature further or adding DMSO. On the plus side, my supervisor is increasingly thinking that the primer is just rubbish and considering getting a new one, or at least letting me do a SYBRgreen assay to determine efficiency ![]()
Hi everyone,
Bad news, we bought a different primer and the results are the same! Any more ideas? I've also tried a different mastermix, but no improvement ![]()