Genotyping by allele specific PCR - (Jan/05/2012 )
hello everyone,
I am trying to genotype an SNP using AS-PCR. The problem is that i have titrated Mg2+ and Ta (52 to 57) but i find only Homozygous mutants been amplified.in around 10 cases i havn't found a single hetero or homo wild. I have noticed that homozygous mutants has quite strong primer dimer but the homozygous wild has no band as well as very faint. The condition of primer s are as follows:
buffer 10x - 2ul
Mg2+ - 0ul
dNTP (o.5nM each) - 1ul
Common Primer(.3uM) - 0.3ul
Polymerase (1U) - 0.033ul
water - 15.07ul
Mutant primer(.3uM) - 0.3ul
Wild primer(.3uM) - 0.3ul
cycling param:
1) 95 for 3 min
2) 95 for 30 sec
3) 54 for 30 sec-----> 32 cycles
4) 72 for 30 sec
5) 72 for 5 mins
the primers have been checked on Sequence manipulation suite and has been passed. What might be wrong. I am attaching a pic. Please don't consider WELL 8 onwards as the primer dimer increased with Mg2+ conc. Please follow from Well 1 to 7. arranged as wild1, mutant1, wild2, mutant2 etc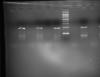
It's odd. Normally it's the opposite that people see non specific amp in AS-PCR, check your common primer design.
The primer design software on NCBI gives Tm as 65.06 and 62.91 for allele specific primers and 59.42 for common primer. By use of thermodynamic parameters available on oligocalc the Nearest Neighbor calculation the primers are as follows 59.94, 58.75 and 55.39. Now the Ta i used was given .. i used gradient of temperature 52,53,55,56,57. Is there sumthn wrong with it??? By sequence manipulation suite all primers are OK and stable.... Pls help ![]()
I concern the complimentary of wt to the common primer more than the annealing temp.
WSN: i couldn't quite get you.. can u be a bit more elaborate and clear???
I meant, even if you have only pair of mutant primer set in reaction, you would get amplification from your wildtypes, unless your wildtypes DNA somehow, 1) not extracted well, or 2) your primers don't recognize your wildtype. Since you have multiple wildtypes that all failed, I suspect it's your primers' problem, morelikely your 'common' primer isn't common to your wildtype. Hope it help!
Looks like something is wrong with your wild type primer. I would check the 3' end to see if the base matches the SNP I am looking for.
