PAGE for Separation of DNA - (Nov/17/2011 )
Hi all,
I have not really used PAGE except for a few basic classes during my under grad.
I have to run some PCR products which lie in the range of 50-300 bp.
We need to do two things
1. Get the best resolution for these PCR products
2. Estimate the band sizes as accurately as possible.
Most labs I know use ABI's Genetic Analyzer for Fragment analysis. However, we just cannot afford the equipment and I intend to use PAGE.
It would be great if you guys could help me with some basics like what is the best concentration for resolving these PCR products, good ladders for estimation of band sizes etc.
Thanks for you help people.
Ameya
Ameya,
I have done this in the past and the resolution of the PAGE is really good, however if you increase the percentage of agarose to 2-3% I think you won't be much worse off. The PAGE will not be a substitute for the chromatographic separation.
Anyway the polyacrylamide percentage for the sizes you specified should be around 12-15%. Contrary to protein electrophoresis you only make one layer of gel, so there is no stacking layer. And you shouldn't add EthBr directly to the gel because it supposedly prevents polymerization. I would also recommend using sodium borate or lithium borate buffer, because the electrophoresis takes longer and generates more heat.
Concerning the ladder, I used Fermentas Low Range DNA Ladder.
Hope it helps.
Miha
Hi Miha and thanks for replying.
We regularly use TBE buffer in the lab. So to start off, I guess we will use TBE. (I do want to try out the SB buffer for a while now). But thanks for the suggestion. I did read about not adding EtBr directly to the gel. Regarding the 2-3% agarose gel, did you mean that 2-3% agarose gel should work well for this purpose. We use 2.5% gels in the lab. But, with this set of experiments we would like to estimate the number of repeats for the loci of the STR. We want to know how accurately can you do that. Therefore, I am presuming that PAGE is a better method to do so.
The concentration of gel that is suggested for the size range I will be working in is variable between 5-15%. But, no one really speaks of the ratio of acrylamide: bisacrylamide here.Is it 29:1 or 37.5:1? Any other suggestions are welcome. Some tips and tricks and beware notices will also help.
Thanks once again.
Ameya
Ameya,
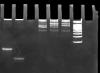
I am attaching a really poor example of a DNA PAGE I did a long time ago. Don't know why but I couldn't find anything better. From the image you can estimate what sort of resolution to expect. I used a 29:1 acrylamide: bisacrylamide ratio and stained with SB buffer with 0.5% EthBr for about 30 min afterwards.
Best of luck.
Miha