Native PAGE - (Oct/24/2011 )
I tried to reverse the electrodes but at that time the pH of buffers was lower than the pI but still near the pI as you said. Then no protein band was observed. Anyway, I'm afraid that by reversing the electrodes, samples totally diffuse.
So maybe next time, I will try to run at acidic pH gel. However, could you please suggest me some possible pH value which suitable for my protein? I just tend to carry out this experiment at the pH 6.5.
you could use the formulation for neutral pH native page posted in this topic. for your purpose i would omit the stacking gel and reverse the electrodes.
i still think that you should use the acid pH native page formulation to which i gave the link in the biotechniques forum.
The other issue here is why you are using 8M Urea for purification? I understand that not all proteins are totally denatured in 8M Urea, but the vast majority of proteins will be partially or completely denatured in a buffer with that concentration of Urea. If your sample is already denatured, why are you running native PAGE? If you need to preserve activity for some downstream step, the 8M urea will probably render your protein unusable unless you do a dialysis/refolding. If you provide more details as to why you are running a native gel vs denaturing gel, we might be able to make alternative suggestions.
Best of Luck.
Thank mdfenko for giving me some suggestions and solutions. I just realized that you're the one who reply me in both 2 forum I've joined in. Btw, many thanks to you ![]()
@Allynspear: The sample containing 8M urea is just as a marker here. Because I didn't run the native gel with the marker. However, I just wonder whether this had any effects on my electrophoresis result. About my sample, after purification, I dialyzed to decrease urea gradually. Thus, my sample now doesn't contain urea anymore.
In addition, my active form of this protein is dimer. That's the reason I run a native gel to observe the dimer band. However, as I said before, I tried some ways but no band was observed.
For that reason, I suppose that I should dialysed one more time under another conditions. But if anyone has better solution, please let me know. Thank you so much!
Observing dimers by gel electrophoresis is not as simple as it may seem from the literature. I myself have tried to do this many times and had great difficulty doing so. I know others who have had similar experiences, and unfortunately there is no single answer. Every protein is different and every protein-protein interaction has different stability. I can tell you that by treating your samples with 8M urea, you are definitely disrupting the dimer and probaly disrupting the folding. In dialysis, you are asking the protein to slowly refold properly and then reform the dimer. This is not impossible, but you are definitely not making it easy for the dimer to form. If there is any way to avoid the denaturation with Urea, you will have a better chance of keeping the dimer intact, or at least keeping a higher percentage of your protein in dimer form. Maintaining native conditions throughout your purification will give you a better chance of observing what you are trying to see.
The other suggestion that I would make is to look into chemical crosslinking of your dimers. There are a ton of different methods out there and it can be very confusing at first. It will likely require optimization to obtain an appropriate amount of crosslinking without going overboard. The major advantage to this is that once the crosslinks have formed, you can perform a lot of different types of analysis without worrying about the dimer falling apart, including SDS-PAGE, which would help with your Native-PAGE problem. I would recommend that you look into some literature about chemical crosslinking, but I might recommend that you look at Pierce's in vitro photocrosslinking system: https://www.piercenet.com/Objects/View.cfm?type=ProductFamily&id=D74D0D83-4A0A-4FB0-BAFF-779654D40043
I don't know what system you are working with, but if you are in cell culture or in vitro expression, the photo-crosslinking system may work for you. If you want, you can provide more detail about your expression system/protein source and I may be able to recommend some options.
Best of Luck.
Hi everyone!
I cannot successfully separate small 8 kDa proteins with native PAGE.
Have tried with BN-PAGE, but there is smear allways present even in presence of detergent dodecyl-maltoside (DDM).
Have already tried gradient BioRad and self made gels (Tris-glycine 10-15-20% gels).
On my gels, higher molecular bands separate nicely, but not small ones.
Does anybody have any tip how to improve separation and reduce smear. Which gels and buffer system so use to separate small proteins?
Tris or tricine?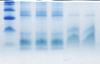

tris-tricine, instead of tris-glycine, is better for small proteins and peptides.
a gradient and/or stacking gel will also help sharpen the bands.
smearing may be caused by aggregation. you should clarify your samples before loading.
