Strange band in colony PCR - (Sep/12/2011 )
Hi,
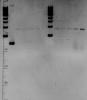
I am quite new in gene cloning and expression. For my experiment, I am trying to clone and express two genes (approx. 1kbp and 1.3kbp) using the pET21a plasmid. I amplify the two genes using PCR and digest the genes and pET21a plasmid using BamHI and XhoI. The RE products are then gel-purify as I got two bands for my PCR products. After that, I ligate and transformed the genes into TOP10 cells. To find my insert, I do colony PCR and got bands (approx. 900bp) for both genes (instead of expected 1.1kbp and 1.5kbp). This is strange as my positive control using empty pET21a vector shows 200bp band. So, thereotically, I should only get band size of 200bp or 1.1/1.5kbp.
Sequencing the clone give sequences outside of the RE site. I would like to know if anyone can give any kind of explanation etc for this. Thanks.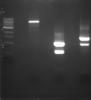
How much sequence are you cutting out of the vector when you do your double digest?
Are you sure your PCR products are what you think they are when you do the gel purification?
Why don't you gel purify the PCR before doing the digest?
Cutting out about 40bp sequence from the vector.
I didn't gel-purify the pcr product as I think i may lose too much product before digestion. I am doing sequencing of the my gel-purified digested inserts to check that the sequence is correct.
