pUC4K HincII sites, how many? - (Aug/28/2011 )
Hello,
I am trying to cut out the kanamycin cassette from plasmid pUC4K with HincII. I have downloaded a corresponding sequence from Genbank and using VectorNTI, I can only see two HincII sites upstream and dowstream from the cassette, wich makes the enzyme perfect for my purpuse. Nevertheless, after migrating on gel I repeatedly get 3 bands rather than two. So does pUC4K has 2 or 3 HincII sites?
Thanks
-
pUC4k should only have two HincII sites! ...what is the size of the bands you see? have you ever loaded uncut vector next to your digest vector? ...maybe one of your bands corresponds to the supercoiled form and your vector is only digest partially? ...if so you'll have to pronlong incubation or increase enzyme concentration.
Regards,
p
I'll do that tomorrow to rule out partial digestion ... and then I'll update the thread.
thx
Plasmid digested, but didn't have to migrate it today ... I'll migrate it early in the morning tomorrow (european time) we'll see ...
Baroudeur
and the long awaited image ... pUC4K has an undescribed HincII site right?? the only other explanation might be that I'm using a mislabled plasmid ...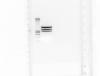
Yes, it looks like it. The only other possible explanation would be uncut plasmid, which is why pDNA suggested running undigested plasmid along with your digestion. Is the sum of the fragment lengths the expected length of your plasmid? Can you cut your plasmid with a single site cutter?
what i meant was put uncut vector next to digested vector on the same agarose gel ...then you would see if one of the bands corresponds to one of the uncut bands.
Regards,
p
yeah guys I know, yesterday I had the chance to run the uncut vector alone but today I forgot to run it along the digest, the drawback of doing many things in parallel. Anyways I'll try to find yesterday's image, and I tend to think this isn't really uncut vector because I gave the enzyme enough time to cut and used 1µ
when i look closely at your pic i see a ~1000 bp, a ~1500 bp and a ~2000 bp band ...to sum up 4500 bp ...pUC4K should be around 3900 bp ...the original bands should be around 1200 bp and 2700 bp ...this corresponds to the two upper bands, so it seems you have some 1000 bp additionally... possibly something is wrong with your vector ...have you checked the identity by digesting with another restriction enzyme? ...maybe it is already pUC4K with some insert?
Regards,
p
P, this is what I suspect: that the petri dish where I'm subculturing the vector from might be mislabled. Have you digested pUC4K with HincII before?
