problem about pIRES2-DsRed2 double digestion - (Jul/19/2010 )
Dear everyone,
I have a problem about pIRES2-DsRed2 (5.3kb,clotech) double digestion. I used XhoI and BamHI to digest the pIRES2-DsRed2 and I got two bands, which are both around 2500bp. I should to be only one band. And then I extracted the fragment (5.3kb) from the gel. I ligated this fragment with my insert fragment(700bp, digested with same enzymes from another plasmid). I transformed the bacteria and picked up the colony. I redigested the plasmid again with same enzymes. To my suprise, these two bands (around) appeared again. After that I use same plasmid( pIRES2-DsRed2 )from another lab , I got same phenomenon. I do not know what happened! I am crazy , could you give me some suggestions.
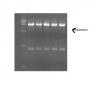
figure 1 lane 1 is the marker ; lane 2 is pIRES2-DsRed2 double digestion with XhoI and BamHI ; lane 3 is another plamid double digestion with XhoI and BamHI . arrow head indicate these bands around 2500bp.
figure2 lane 1 is marker. lane 2-6 is ligated plasmid double digestion with XhoI and BamHI . All of them have my insert fragment.arrow head indicate these bands around 2500bp.![]()
This is probably some minor star activity of one of your enzymes. Don't worry about it. It looks as if your ligations worked. Forward. If it worries you, turn down the exposure time on your gel images.
phage434 on Jul 20 2010, 04:49 AM said:
Dear phage434,
Thanks very much! the enzymes in my double digest buffer have no star activity.However the activity of XhoI in this buffer is 50-100.I really do not know what happened!
sincerely
Tao
