DNA sonication - RNA contamination? - New probe, start all over (Mar/17/2010 )
OK so I am using a new probe with my Branson 450 digital sonicator. Turned out I was using the probe meant for, shall we say, slightly larger volumes ![]() . In any case, I got the correct (weaker) probe and have to start over. Anyway what do you guys reckon of my sonication?
. In any case, I got the correct (weaker) probe and have to start over. Anyway what do you guys reckon of my sonication?
I sonicated for at 25% amplitude using 10 second pulses with 30 seconds rest on ice for:
Lane 1 - 6 rounds
Lane 2 - 10 rounds
Lane 3 - 15 rounds
Lane 4 - 20 rounds
I am using the Magnify Chip kit and purifying the DNA using the magnetic beads with the kit.
Personally I think it looks pretty good, it could maybe do with 25 - 30 rounds but then I think it might be overdone a little. I could also increase the duration of each pulse and probably reduce the rounds, but at some point you have to be happy with what you have. Any comments?
It appears to me there is over sonication because the brightest band is at 100bp while there is barely smear in the 200 -1000 bp range. In addition, I did not see much difference between 6 rounds and 20 rounds. I think 10 second pulse is appropriate.
I'm not used to this sonicator. But how many cells were you starting with? How much lysis buffer?
pcrman on Mar 17 2010, 07:45 PM said:
Yeh I think the low bright band is RNA personally, dont you? The samples are not RNAse treated. That leaves the higher smear to be DNA in my opinion, which appears to range from about 1000 - 200 bp. This is only a very small aliquot of my sample (20 ul) so recovery is not the greatest.
Oh and it is about 1 x 10^7 in 500 ul IP buffer (without SDS)
That's it???????? Almost 100 views and no more comments. Really would like some feedback here guys.
Dukey on Mar 23 2010, 08:04 AM said:
I would agree with pcrman, your samples look over-sonicated. Even if we're wrong, at the very least you need to repeat this with an RNase digestion. Surely there will be a load of RNAs in the 200-1000 bp range that will distort what you're seeing anyway.
looking over the MAGnify manual.. it says to use 1million cells per 50ul Lysis buffer. What's the reason to lyse in IP buffer without SDS?
Thanks
nigelcr on Mar 23 2010, 07:33 AM said:
Dukey on Mar 23 2010, 08:04 AM said:
I would agree with pcrman, your samples look over-sonicated. Even if we're wrong, at the very least you need to repeat this with an RNase digestion. Surely there will be a load of RNAs in the 200-1000 bp range that will distort what you're seeing anyway.
OK I'll entertain the idea that this is RNA and post back when I have performed the RNAse treatment.
OK so here we go. Basically performed the same experiment and then treated an aliquot with RNase to demonstrate that the low band (<200 bp) was indeed RNA. You can clearly see the difference and it is obvious that the sonication is doing the job as there is a nice smear all the way down to 200 bp. I think the experiment highlights how important it is to RNase treat when checking sonication as the RNA can really confuse the interpretation of the gel. I was being lazy and not doing it ![]() I do not really believe you can get DNA <200 bp with sonication and I think this goes some way to proving that.
I do not really believe you can get DNA <200 bp with sonication and I think this goes some way to proving that.
I changed a couple of things with DNA purification as it is far too costly to do all this work using the Magnify Chip Kit. I ethanol precipated 50 ul of my sonication and dissolved the pellet in 100 ul chelex-100, boiled for 10 minutes and then treated with pro-k for 30 minutes at 55 degrees. I Spun down the chelex and took the supernatant and then I re-precipitated again overnight as I wanted to concentrate the DNA for loading and for RNase treatment. I dissolved the pellet in 50 ul of water and treated 25 ul with 2 ul of RNase cocktail (Ambion) for 30 minutes at 37 degrees. Ran gel.
Lane 1 - No sonication
Lane 2 - 5 rounds (20%)
Lane 3 - 10 rounds (20%)
Lane 4 - 15 rounds (20%)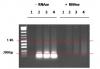
happy to see you got your profile. keep in mind if your are planning on sequencing, that chelex resin generates too much ssDNA...really difficult to get enough dsDNA for library preparation.
