miRNA isolation from serum - (Mar/17/2010 )
Hello community!
I have been searching the forum for a conclusive description of miRNA serum extraction, but I haven't found anything beyond references to "Trizol extraction, the usual, you know". ![]()
I have already tried 2 kits (Ambion miRVana, Qiagen miRNeasy) and for the Qiagen kit I have tried some extra wash steps and also precipitation. I have however never reached sufficient RNA quantities, even when starting with 1ml of serum.
So now my questions:
0) What kit / reagents do you use for totalRNA extraction from serum and what modifications do you apply to the manufacturer's protocol?
1) I tried precipitation with glycogen during extraction, but I never saw anything close to a pellet! Since the original kit instructions (Qiagen, miRNeasy) don't include a precipitation step, I went ahead and proceeded with the entire volume and added it to the column. What effects do you expect when not getting the pellet and therefore not washing it? What could be the reason for my missing pellett?
2) Has anyone ever used serum samples for miRNA microarray?
3) How do you determine your RNA concentration? I was told that neither photometer, nor nanodrop are able to deliver remotely useful data. I've read that some of you spike in C. elegans RNAs to normalize with (which I think is pretty cool), but still: not everyone does this...
4) How much serum do YOU need to produce 1µg of totalRNA?
Thanks for your help and insight, I know that there are partial answers to these questions in the forum and I have read these, but I'd be grateful if a number of people posted their collective experience here so we can get a general picture.
I added a nanodrop bioanalyzer pic of my serum samples. There is a faint band in the miRNA region. These samples were analyzed by microarray. 8 samples in total, derived from 2 biological samples. So in all, we have 2x 4 replicates, but they totally failed to cluster appropriately.

Thanks a lot & cheers!
I don't understand, why do you say Nanodrop is not reliable for RNA? Because of RNase Inhibitor? try measuring before adding such components.
last time I used a kit for mRNA extraction from Roche Applied Science, but it also needed DYNAL magnet racks because the extraction principle relied on magnetic beads. But for miRNA I'm not sure.
I use Qiagen Mini easy for total RNA. it has a red box. Trizol is also good, but I only use Trizol if my sample has a small volume such as virus samples. But for Eukaryotic samples I use Qiagen kit.
Curtis on Mar 17 2010, 07:58 PM said:
last time I used a kit for mRNA extraction from Roche Applied Science, but it also needed DYNAL magnet racks because the extraction principle relied on magnetic beads. But for miRNA I'm not sure.
I use Qiagen Mini easy for total RNA. it has a red box. Trizol is also good, but I only use Trizol if my sample has a small volume such as virus samples. But for Eukaryotic samples I use Qiagen kit.
- I readily admit that I can't give you facts why nanodrop is supposed to not be suitable, its a comment I've come across talking to several people and that I've seen here in the forum, too. Can someone clarify (or correct my feelings about the nanodrop)?
- Don't the Qiagen Mini Kits lose the miRNA fraction? I believe you need the miRNeasy Kits. In any case, it sounds like you are extracting RNA from tissue, not serum? I'm specifically troubled by serum, tissue is working fine so far.
Cheers
They have kits for everything, after all they have the widest range of kits in this business.
I use Nanodrop myself. I have no problem with it.
+1 for the Nanodrop. No problems unless your concentration is really low (<10 ng or so).
For the isolation: Chen et al Cell Research 2008 18:997. Equal vol of Trizol to serum, and 3x P/C extractions, to get 5-10ug RNA from 50 ml serum.
Also check out Mitchell PNAS 2008 105(30):10513. They used an Ambion kit, but I don't currently have access to the supplementary information where the methods are contained.

hi,
First what is your study model ? cancer cells ? which kind ?....
I am working on circulating miRNA as well. there are two schools for excreted miRNA isolation and prior to the trizol or any kit isolation.
Some teams first isolate the exosomes, which is long but you re sure you don t have any contamination !
the other one is direct isolation from serum/culture supernatant.
I am going to do a comparision btw those two. I ll know more next week !
Concerning which kit you should use, the guys from the functional genomic unit in my lab (who do the arrays) told me that Trizol/tri-reagent was mor suitable because you may loose miRNA with the columns, they don t retain all miRNA.
that s why they recommand to use trizol with glycogen.
concerning nanodrop, it is supposed to not be reliable for DNA concentration. try a comparision btw DNA concentration data from nanodrop and a fluorescence technique with an DNA intercalant. We don t know why but there is a difference and it is the kind of detail very important when you run an array ! some late nature news issue claims that plastic tube release some chemical compounds (probably monomers) into you DNA solution and thus interfers with UV absorption...
but it s weird because it works well for RNAs...
cheers.
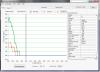
I have very similar problems. After extraction of RNA from plasma (Ambion miRVana kit) I usually got about 10-40 ng/mkl according to Nanodrop, but the 260/280 ratio is always about 1.4 which is not normal for RNA, and 260/230 is awful 0.2-0.4. Upon running on Agilent Bioanalyzer, it seems that there is no RNA in these samples.
I contacted people who work routinely with miRNA from plasma and they told that they also have abnormal 260/230 ratios after columns, and that one cannot use Nanodrop readings for RNA quantitation for these cases.
We have found similar problems using Trizol LS to purify miRNAs from plasma and spent considerable time on this. It is true you do get aberrant results by nanodrop the thing to look out for is the low 230/260 ratio which although indicative of salt contamination is more likely to be caused by phenol contaimination which absorbs very strongly at 270nm (you can see this on the spectrum of the nanodrop). I personally believe that this is a problem caused by the very high protein levels in plasma/serum samples. What we did was to clean up the miRNA in RNeasy columns using hig EtOH concentrations. You aparently 'loose' alot but it probably wasn't really there to start with. You can then see on a Bioanalyzer
Hi.
Just to be up front, I should mention that I work for NanoDrop.
pbadtopo is correct that phenol contamination would be a problem for spectrophotometric measurement of nucleic acid concentration such as using a NanoDrop. Phenol will absorb at the lower end of the spectra (less than 230 nm) and also at around 270 nm. Often if you have phenol contamination, you will look at the your concentration reading and 260/280 ratio and think that you have lots of nucleic acid and that it is nice and pure. If you look closely, you will be able to tell this is not so, and here's how:
Look at the 260/230 ratio - this will usually be poor.
Look at the trough in the spectra - this should be at 230 nm. If it is at a higher wavelength (e.g. more than 235 nm) then you have some contaminant (not necessarily phenol).
Look at the peak in the spectra - this should be at 260 nm. A peak at 270 nm or thereabouts is fairly characteristic of phenol contamination.
Once you have used the spectra to discover that you have a problem, consider both your starting material and your extraction method.
If you get unusual spectra or have questions about contaminant detection, please call or email us (number and address are on the NanoDrop website) - our tech team has a ton of experience with this and can look at your NanoDrop data and help.
Cheers.
well, Limey, just to mention - on the "Contact Technical Support" page there is no way to attach data, and no email address (the only address displayed is on common "Contact Us" page).
in my case, spectra look really quite strange (see attached). First, they are kind of serrated; second - there is nothing for wavelengths after 250 nm, although A260 number is not zero.