Contamination in Multilplex? - (Mar/15/2010 )
Hi to all. Hope some Masters could help me out with my problem which has
taking place for 2-3 months.
I currenty conducting a research on a reptile species genetic variation using microsat primers/markers (cross-amplified from a close related species). For this while, I have been "haunted" with "contamination" alike problem. The NTC (negative control) in the multiplex (TouchDown PCR with 35 cycles, all primers are flourescent labelled primers (reversed one); blue (6fam), yellow (ned), red (pet), green (vic)) seems keep showing band/s on and off, particularly the NED labelleds. At first I thought the primers aliquot or the reagents might be contaminated and I've had changed all reagants and have made new aliquots (always work with Lamina Flow, with autoclaved tips, water, etc, I aeven have asked few labmates to set up the PCR for me using their reagents, the same problem still persist). I have genotyped the problematic NTC, it shows there is a microsat peak/s in the NTC with size (157bp) that close to my wanted product size (ca 150-160bp), image attached. But this microsat peak/s has never found in my other templates. This is very strange that if one or more of my primers were contaminated the contamination peak/s should have had appeared in my other templates (because it's mutliplex) other than in my NTC, and other non-contaminated primers in the multiplex would have also amplified the contaminant eventually more different peaks of different coloured labelled peaks would have showed up too in gel or genotyping. I have tried to run them individully on singleplex and they were look good and yet with less messy and stuttery.
Pls see my photo which shows only the NED laballed "contamination" peak found in the NTC and it does not show up in other templates t07025 etc
Could anyone enlighten me? Cheers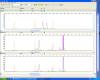
The first thing I would do is to stop using autoclaved tips for PCR and change to RNase/DNase-free filter tips. The same with the water, do not autoclave water but use ultrapure water like this https://products.invitrogen.com/ivgn/produc...=Search-Product for PCR and all associated procedures like primer dilutions, cDNA reactions etc.
Thanks. I am actually using the tips you mentioned and the water is Sterile MilliQ water. BTW, does it is really a contamination which taking place? Very weird as only one product peak is being shown.

i think it is " how you deal with your NTC " ...
i, suffered from the same .. somehow !
and what i have done, is changing the sequence for the addition of the positive control/my samples..
bring two racks in the hood,
put the empty tubes in the first rack and add the mix in all.
then,
add water to ur NTC and transfer it to the second rack, then the negative control and transfer it ...etc
being the last tube the positive control ...
change ur gloves meanwhile, and keep soaking ur hands and the racks with bleach 10% ...
hope my tips will aid.
Have you solve the problem?
I don't think it's contamination,maybe PDs ……
What about sequencing the unwanted product?
good luck ![]()
![]()
