Removal of RNA from DNA sample - (Nov/20/2009 )
Dear all
I'm really unhappy with my DNA isolation at this moment since I cannot get ride of RNA from the sample, which I've heard will interfere with DNA during PCR. For removal of RNA I did RNAse treatment and then phenol extraction and CIA (chloroform+isoamyl alcohol) extraction. The DNA was precipitated using NaOAC and absolute ethanol, pelleted, washed with 70% ice-cold ethanol, dissolved in TE solution and the concentration was measured using Nanodrop. The ration 260:280 is all the time around 2. I repeated RNase treatment in same sample but no improve.
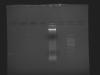
To look for the presence of other stuffs along with DNA, I also did ran agarose gel with the sample. Here is also the photo of the gel (sample and ladder). It seems from gel my DNA is not pure. I require some help, please!
The Question on Nov 20 2009, 12:38 PM said:
I'm really unhappy with my DNA isolation at this moment since I cannot get ride of RNA from the sample, which I've heard will interfere with DNA during PCR. For removal of RNA I did RNAse treatment and then phenol extraction and CIA (chloroform+isoamyl alcohol) extraction. The DNA was precipitated using NaOAC and absolute ethanol, pelleted, washed with 70% ice-cold ethanol, dissolved in TE solution and the concentration was measured using Nanodrop. The ration 260:280 is all the time around 2. I repeated RNase treatment in same sample but no improve.
To look for the presence of other stuffs along with DNA, I also did ran agarose gel with the sample. Here is also the photo of the gel (sample and ladder). It seems from gel my DNA is not pure. I require some help, please!
Have you tried to do PCR on your DNA/RNA extract and checked whether possible RNA interferes with your reaction? Usually it's the other way around, that's why many automated DNA extraction protocols don't have an extra RNase step. I would have tried the PCR just to give it a check.
The ususal problem with nanodrop measurements on DNA samples containing RNA is that is measures both, which again may give you a false high "DNA" concentration and a too rough dilution of the DNA afterwards. Have you tried measuring DNA by picogreen or other fluorescence methods?
DocFlow on Nov 20 2009, 01:00 PM said:
The Question on Nov 20 2009, 12:38 PM said:
I'm really unhappy with my DNA isolation at this moment since I cannot get ride of RNA from the sample, which I've heard will interfere with DNA during PCR. For removal of RNA I did RNAse treatment and then phenol extraction and CIA (chloroform+isoamyl alcohol) extraction. The DNA was precipitated using NaOAC and absolute ethanol, pelleted, washed with 70% ice-cold ethanol, dissolved in TE solution and the concentration was measured using Nanodrop. The ration 260:280 is all the time around 2. I repeated RNase treatment in same sample but no improve.
To look for the presence of other stuffs along with DNA, I also did ran agarose gel with the sample. Here is also the photo of the gel (sample and ladder). It seems from gel my DNA is not pure. I require some help, please!
Have you tried to do PCR on your DNA/RNA extract and checked whether possible RNA interferes with your reaction? Usually it's the other way around, that's why many automated DNA extraction protocols don't have an extra RNase step. I would have tried the PCR just to give it a check.
The ususal problem with nanodrop measurements on DNA samples containing RNA is that is measures both, which again may give you a false high "DNA" concentration and a too rough dilution of the DNA afterwards. Have you tried measuring DNA by picogreen or other fluorescence methods?
Thanks for reply
I used isolated DNA for qPCR. I get the required product. But the problem is I don't get efficient result. My supervisor was saying for the preparation of standard curve with isolated DNA first instead of plasmid DNA. The low efficiency of my qPCR result could be due to the presence of RNA in my sample, I think.
What if you physically isolate/cut out the correct band from your gel and do another PCR on it? If you still see the additional band and smear on the next agarose gel the problem might be insufficient PCR reactions and not interfering RNA.
DocFlow on Nov 23 2009, 09:36 AM said:
Hi DocFlow
Thanks again for reply. That could be something I still can try. By the way the gel I've shown above is just from isolated DNA but not from PCR product.
hi TQ, Can you tell us what was the method you used for DNA isolation? What was your starting material, cells/tissue??
can u elaborate what do u mean when u say low effficiency of the PCR?? are u saying sensitivity is complromised? or hight Ct values?
Are u doing a Q-PCR?.. as doc said... rna in your sample will not effect the amplification reaction most of the times.
Pradeep Iyer on Nov 23 2009, 11:54 AM said:
Are u doing a Q-PCR?.. as doc said... rna in your sample will not effect the amplification reaction most of the times.
I used phenol extraction method for C elegans. I meant indeed there could be some problem in sensitivity due to the presence of RNA in the sample. I heard from my supervisor as well low sensitivity could be caused due to the presence of high amount of RNA as well. In my qPCR for instance, sometimes I get higher Ct values for lower dilutions.
well ya tat means your RNA might be interfering with the reactions.. or alternatively due to the presense of RNA you are quantifying the DNA wrongly (over estimating it) and hence adding less amount than what shud be added so the lower sensitivity!!
So at hgher dilutions that is more concentration the Ct values are unaffected??
How does your positive control behave???
Pradeep Iyer on Nov 23 2009, 12:10 PM said:
So at hgher dilutions that is more concentration the Ct values are unaffected??
How does your positive control behave???
I don't know why I get slightly higher or similar Ct values for higher dilutions (compared to lower dilution). And at this point I was thinking RNA as a probable problem. What do you mean by positive control here? I was willing to use isolated DNA to make my standard curve instead of plasmid DNA. So, I used isolated DNA and negative control for my qPCR.
