SOS....Genomic DNA extraction - Extraction from gram positive bacillus stearothermophilus (Oct/09/2009 )
Hi, everyone...
My name is Goh... Nice to meet u guys in these forum...
I have started my Master project 2 months ago and faced problem in extracting genomic DNA from bacteria of interest, Bacillus stearothermophilus. I failed to isolate the genomic DNA from this gram positive bacteria. It was most possible due to the problem of cell wall lysis.
I have used 20mg/mL lysozyme (in solution contained 20mMTris-HCl, 2.5mM EDTA, 1% triton X-100) to lyse the cells with incubation times of 30, 60 and 120 minutes respectively....I found that the solution has turned to clear but got a lot of white precipitates on the bottom of tube...Hence, I am not sure whether the cell lysis is completed or not?????
Recently, I found that there is a few of lytic enzyme (besides of lysozyme) can be used to effiently lyse the gram positive cells.... such as mutanolysin, lysostaphin, and libiase.... Have anyone could tell me which one is better and cost effective used for breaking of cell wall of gram positive bacteria???
Look forward for a reply soon....
Thanks a lot... Have a nice day...
Try this:
1. 50ml suspension with OD730=1.2-2;
2. 3krpm, 4C, 10';
3. Get the cells from the bottom.
4. wash in 1.5ml Tris/EDTA buffer;
5. 15krpm, 4C, 1';
6. pour out liquid phase, add 270Ál STET, shake;
7. add 30Ál lysozyme (50mg/ml);
8. 37oC, 30';
9. add 100Ál 10% SDS;*
10. 65oC, 40';*
11. add 100Ál 5М NaCl;
12. 65oC, 20';
13. add ~200Ál 1М NaCl (if volume is not enough);*
14. add 1V chloroform and shake 3'
15. 10krpm, NT, 10';
16. Get the upper phase and goto 14 (one or two times*)
17. Add 1V isopropanol
18. 4C, 15-20';
19. 15krpm, 4oC, 10'
20. wash 70% ethanol;
21. delute in 50Ál H2O.
STET:
(store +4C)
8% sucrose
5% triton Х-100
50mМ EDTA-Na2, рН=8.0,
50mM Tris-HCl, рН=8.0.
Tris/EDTA:
(store +4C)
50mМ Tris-HCl, рН=8.0
50mМ EDTA-Na2, рН=8.0
*Optional stages.
Dear gleb. kudr,
Thank you very much for the method that you have provided to me... I will try it... ![]()
Besides, may I know that after lysis step, could I continue purify the genomic DNA by using commercial genomic DNA Purification Kit???
goh on Oct 9 2009, 05:44 PM said:
Yes.
From step 14 where you added chlorochorm you may use purification kit instead.
Dear gleb. kudr,
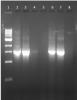
Hi, I have tried the methods you gave me... I purified the DNA by using commercial kit instead of chloroform and added proteinase K after step 14 ... after gel ectrophoresis analysis of my extraction products, i got a very thick band/smear that lower tha 1000bp.... I wonder that is it the extraction products was RNAs or the fragments of genomic DNA? I think mostly is my genomic DNA was degraded...
May I know why my genomic DNA was degraded? Is it related to the addition of proteinase K?
Besides, could I know what's the function of NaCl in the method that you have provided me..?
Thank you very much....
U're supposed to be seeing something at like 10,000bp and above size range for successful gDNA. if u think 1000bp is RNA, u can treat it with RNAse .
NaCl helps to neutralize or shell the negatively charged phosphate bone of nucleic acid to faciliate its precipitation.
U can actually go on column purification straight away as long as u did a lysozyme treatment beforehand.
increasing the lysis incubation time with occasional vortex or constant slow rocking will improve yield.
what kit are u using?
which Uni? UKM?UPM?USM?UM?UTM?
Dear Hanming,
Thanks for ur suggestion.... I ever tried breaking the cells with only lysozyme (20mg/mL) and then straight away continue with purification kit...solution turned clear after 1 to 2 hours incubation...but got lot of whitish precipitates at the bottom of tubes... then is it the lysis process is completed??? And I couldnt get any dna from it...
I used promega and biobasic purification kit.... got any recommendation?? especially for dna isolation from gram positive bacteria...
I came from UM...
Thank you...![]()
u can try 2 thigns
1.decrease cell mass used for DNA purification
2. increase lysis time ( extend till O/N)
both in combination won't hurt as well.
show us a picture of the gel if possible.
promega is an OK kit assuming everything is still fresh and intact.a good lysis won't have anything left. the white precipitate somewhat indicate incomplete lysis sometimes.might just be spheroplast or something
It's RNA. Use rnase.
