Primer Dimer in NTC Only? - Primers Dimers show up in NTC but not template wells (Sep/12/2009 )
I have a question concerning primer dimers; I have not been able to find an answer to this yet in the forums:
Background: I've been starting up qPCR in our lab, we have an ABI 7500 fast system and I'm using the Fast SYBR Green Master Mix from ABI. I'm working on determining primer efficiencies for primer sets to be used to analyze chromatin immunoprecipitation data. The amplicon size for the primers range from 250-350 bps and are tiled along the presumed promoter region of our gene of interest.
Issue: I've examined efficiencies for some of my primer sets and they come out quite good (around 95%), however, I seem to get amplification in my NTC wells that appear to be primer dimers. In my template wells (mouse gDNA) at 1:1. 1:10 and 1:100 dilutions I get very nice amplification, clean melt curves (no shoulder) and a nice solid band on my gel. However, in my NTC wells (typically 2/3) I also get amplification that is not my product and has a lower Tm. Upon running on a gel it appears as a diffuse band below 100bps (looks like a primer dimer, smells like a primer dimer...).
Question: Is this an issue for experimental analysis? Can something like this be solved simply by playing with primer concentrations? Or does it require primer re-design? Would switching master mixes potentially help (e.g, to Power SYBR Green)? I'm a bit limited in my genome region as I need to stick around my presumed promoter region for the ChIP.
Thanks for considering my question.
You probably need to do a little more work to optimize your cycle conditions for your primers. First, I would recommend doing Gradient PCR and use the highest annealing temp that gives you a he correct size-specific product, without pd formation. Higher temperatures can also improve your efficiency since you increase specificity. Alternatively, you could also use a lower primer concentration, but you might still detect pd fluorescence in your late cycles
Thanks for the response jah. I have to admit I'm a bit of an amateur at qPCR and our lab doesn't have much expertise. I've gone back and taken a closer look at my primers using Net Primer and realize my problem is likely the formation of cross dimers. Your suggestion sounds good though, in my primer sets in which I only seem to have a small primer-dimer problem I'll try playing with the annealing temperature. I've just been using a 2-step PCR as recommended by ABI for their master mix (95C and 60C) and hoping that will give me the best results.

Mighty Mouse on Sep 17 2009, 05:27 PM said:
I agree with Jah, you need to play with annealing temperature.
And you can give me your primers, I'll check it in Vector NTI.
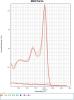
Thanks for your responses thus far...so I've got a question regarding the interpretation of the melt curve data. There are some of my primers that come out beautifully, however I'm not sure how to interpret the melt curve data from other primer sets (see attached). It seems that I have two outcomes that indicate primer dimers. I'm wondering if one particular outcome versus the other would suggest a particular solution for overcoming the primer dimer issue.
All the figures below represent the derivative output from the melt curve for NTC and mouse gDNA samples.
Primer 8(1) seems to produce a very clear primer dimer at a Tm immediately prior to the expected amplicon.
Primer 58(1) has a broad "shoulder" prior to the expected amplicon.
Interestingly, in both sets there does not seem to be any primer dimer amplification in the NTCs. On the gel for primer 8(1) there is clearly only one band of the appropriate size. My gel with 58(1) didn't come out well enough for me to get a good look at it, but it basically looks good in terms of specificity.
Would increasing the annealing temperature work better for one particular melt curve outcome versus another?
Thanks!