PCR problem - (Sep/08/2009 )
Hey All, I've got a really "nasty" target to amplify...
1. cDNA sequence is clear.
2. gDNA (2.2kb) has 6 exons with a gap (~600bp) between the 4th and 6th exon on chromosome A.
3. The newly published genome data is different from the previous version. The whole gene is spliced: exon 6 is located onto another scaffold/chromosome while exon 4 is lost in the entire new-version. And I also see big rearrangement on the chromosome A.
4. We've got three different person working on this gene...no success yet.
5. What we got: I'm able to span the first 3 exons and 2 introns, as well as the last part of this gene (exon 6 and 3'UTR), but was never able to span the gap region... If anything showing on the gel, the band size corresponds to the amplicon using cDNA as template.....though I'm sure I'm using gDNA, and no RNA/cDNA contamination..
6. I tried different optimization strategies, none had work so far. (Tm, extension time, denature time, Mg conc, enzyme digestion, different polymerases, deaza-dGTP...)
I hope I could describe this problem more clearly.....sorry in advance ~~
My question is could that be a real trans-splice? gDNA from different chromosome, spliced together to make a real cDNA? OR, is that true there is no intron at all, although the sequence information from genome database shows there must be 1-2 introns..OR, this gene is in a very "fragile" region of a chromosome, which splits easily during DNA preparation?
And how to prove/disprove for each case?
I'm in desperate now..help me out ~~~ ![]() ~~~~
~~~~
have you tried to amplify smaller parts of that intron?
have you looked to see if there are things like stem loops in there?
have you tried using additatives, like DMSO et al?
have you tried different primers for that region?
vetticus3 on Sep 9 2009, 04:03 PM said:
have you looked to see if there are things like stem loops in there?
have you tried using additatives, like DMSO et al?
have you tried different primers for that region?
Hi Vetticus, thanks for replying~
My answers:1.... I was able to amplify the first 3 exons and 2 introns, as well as the last part of this gene (exon 6 and 3'UTR), but was never able to span the gap region..including the intron in that gap..
2.. gDNA sequence is not available in that gap...don't know how to look at the stem loop in that region..could you give me more details?
3..yes...not worked
4..yes...3kb thing..i designed 21+ primers..all the primers in the known region worked..but never able to span the gap..
so for example, I have region A, B, C. A and C are known region and B is the gap region. I designed my primer in the B region based on cDNA sequence.
In A, I have primer called Af (forward), and Ar (reverse); In B, I have Bf, Br; In C, I have Cf, Cr.
So, A and C are known region and B is the gap region. I designed my primer in the B region based on cDNA sequence.
Af-Ar, PCR positive, size right;
Bf-Br, PCR positive, size=amplicon from cDNA template;
Cf-Cr, PCR positive, size right..
The question is never able to amplify A to B, A to C, B to C....if anything showing on the gel, the size corresponds to the thing amplified from cDNA template....
Hope I explained that clear~~
Thanks again~
if you don't know the sequence, how did you design primers for that gap?
what i would do is take the primers that worked, which are closest to that gap, and use them in a sequencing reaction.
V
vetticus3 on Sep 10 2009, 03:37 PM said:
what i would do is take the primers that worked, which are closest to that gap, and use them in a sequencing reaction.
V
Sorry V, I didn't make it clear. We know the whole cDNA sequence, but we have no clue about the gDNA sequence for that gap. cDNA sequence shows there is one exon inside that gap, so I basically designed primers based on the exon sequence in the gap.
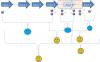
I've loaded a picture hope I can explain better this time.
So I have 6 exons, with a exon-5 in the gap region;
Primer 1, 2 works (indicated by smile...), which should mean its reverse primer 3 is good.
Primer 4,6 works, which also should mean their reverse primer 5, 7 is good.
Primer 8,0 works, which also should mean its reverse primer 9 is good.
But, primer 3, 5 never worked (indicated by
.....
Hope you can understand me this time~~~