Fragmented DNA doesn't appear as bands on agarose gel - (Jun/17/2009 )
Hello all
we face a problem concerning qualitative analysis of fragmented DNA
we extract DNA from liver of mouse and load the DNA sample on agarose gel but we didn't detect clear band.
photo is attached.
PLEASE Helppppppppppppppppp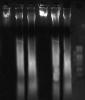
-jehane-
What did you exactly do? Did you extract DNA and load it onto the gel directly?
-dnafactory-
dnafactory on Jun 17 2009, 03:39 AM said:
What did you exactly do? Did you extract DNA and load it onto the gel directly?
at first homogenize liver tissue on TES lysis buffer
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
-jehane-
jehane on Jun 17 2009, 12:45 PM said:
at first homogenize liver tissue on TES lysis buffer
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
Than, this is the correct image of your DNA. You see a smear when you treat this way. If you use DNase or you sonicate, you will see some bands...
-dnafactory-
dnafactory on Jun 17 2009, 04:09 AM said:
jehane on Jun 17 2009, 12:45 PM said:
at first homogenize liver tissue on TES lysis buffer
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
Than, this is the correct image of your DNA. You see a smear when you treat this way. If you use DNase or you sonicate, you will see some bands...
Using DNase or RNase??
sonication will affect the fragmentation?????????????????/
-jehane-
jehane on Jun 19 2009, 04:53 PM said:
dnafactory on Jun 17 2009, 04:09 AM said:
jehane on Jun 17 2009, 12:45 PM said:
at first homogenize liver tissue on TES lysis buffer
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
then take 100 ul of the homogenate to new eppendorf adding 50 ul proteinase k - incubate at 37 C with shaking overnight
then load sample onto the gel
Than, this is the correct image of your DNA. You see a smear when you treat this way. If you use DNase or you sonicate, you will see some bands...
Using DNase or RNase??
sonication will affect the fragmentation?????????????????/
Well, when you sonicate you shear the DNA and you areable to see distinct bands. If you do not sonicate, you usually see a lot of DNA in the well and a smear. Using micrococcus DNase will have the same effect of sonication.
As for "normal" images of DNA you can have a look here: www.biomedcentral.com/1471-2202/6/13/figure/F2
-dnafactory-
