DNase 1 treratment of RNA - (May/15/2010 )
Dear friends,
Has anyone used DNase 1 ( fermentas) for removal of genomic DNA from RNA preparations. I tretated my RNA preparation with dnase 1 according to the protocol ut the RNA got degraded.
My protocol was as follows:
RNA: 1 microgram
10 X DNase 1 buffer with MgCl2: 1 ul
DEPC treated water : 9ul
DNase 1 (1U/ul) : 1 ul
Incubate at 37 degrees for 30 min.
add 2 ul of 0.5M EDTA to it and heat at 65 degrees to inactivate DNase 1
but after doing this the DNase1 treated RNA is degraded as seen on formaldehyde agarose gel.
Does heat inactivation (65 degrees , 5 min) cause RNA to degrade?. or else is there any other option?
pls help
it is possible that your DNase is contaminated with RNase (since RNase's are very stable proteins) ...but i would not suggest that this is your problem.
What makes you think that your RNA is degraded? How do you measured the RNA concentration before the digest? Did you checked it by gel electrophoresis? ...or how do you differentiate between DNA and RNA?
Regards,
p
I agree with pDNA that RNase such as RNaseA and RNaseT1 are very stable, and they cut RNA very quickly. Have you used the RNase Inhibitor (or RNaseIn or RNAguard) in your experiment? Without protection, even chase amount of conteminated RNases will cause your RNA degradation. Make sure your RNA is pure enough, and make sure the DNase I and EDTA you used is certified to be RNase free. Finally, always add the RNase Inhibitor or something like that in the reaction mixture to protect your RNA.
i checked the quality of RNA on a formaldehyde agarose gel after the DNase 1 treatment. The rRNA bands appeared smeared and not intact. also the RT-PCR did not work.
Dear zhongmindai
I did not add RNase inhibitor during the dnase 1 treatment of RNA. I shall try adding the rnasin inhibitor to the RNA prep when DNase 1 tretament is given. I have a few doubts
1. Is the RNase inhibitor heat stable?
2. Will addition of RNase inhibitor affect the Reverse transcription step of the RT-PCR.
3. Is it necessary to add rnase inhibitor during the RT-PCR step.
pls help
chn09 on May 16 2010, 09:05 PM said:
I did not add RNase inhibitor during the dnase 1 treatment of RNA. I shall try adding the rnasin inhibitor to the RNA prep when DNase 1 tretament is given. I have a few doubts
1. Is the RNase inhibitor heat stable?
2. Will addition of RNase inhibitor affect the Reverse transcription step of the RT-PCR.
3. Is it necessary to add rnase inhibitor during the RT-PCR step.
pls help
Most RNase Inhibitor is not heat stable (a company said their kind of RNase inhibitor-like products can work at higher temperature such as 50 oC, but I forget its commercial name).
Addition of Rnase Inhibitor prevent the RNA from degradation during reverse transcription. We always add the RNase Inhibitor in our reverse transcription reactions. RNase inhibitor is necessary unless all your reaction mixtures are rnase free.
Have you checked the RNA quality before treating it with DNAse? Maybe it has degraded beforehand. Also, make sure your DNAse, buffer, and EDTA are all RNAse-free. I use the DNAse from Promega (comes with buffer and stop solution), and it works just fine. The heating step should not degrade the RNA.
Lena
Chn09,
I am afraid that you had RNase contamination, either it was in your RNA prep or in the DNase I. It's always a good idea to test if your RNA prep is contaminated with RNases before using it in downstream work.
To test for RNase contamination, you could take an aliquot of your RNA prep, add DNase buffer to 1X, divide it into two, add DNase I to one, incubate both at RT for 30 min, and then run the gel.
If you see RNA degradation in both buffer only and DNase lanes, your RNA is likely contaminated (most likely). If you see RNA degradation in DNase lane only, your DNase I is contaminated (sometimes, from some vendors).
Good luck,
aquaplasmid
"Finally, total RNA can be extracted from frozen whole blood!"
Hello,
I am not using DNase 1 to treat my RNA but RQ1 RNase-Free DNase from Promega. There is a bit in the protocol that comes with the enzyme that states that the reaction buffer included with the enzyme effects gel electrophoresis. The buffer apparently causes aberrant migration and smearing of RNA on gels. Is it possible you RNA is in tact and the gel is just being affected by the reaction buffer you used for your DNase treatment? Promega recommend carrying out a phenol:chloroform extraction and ethanol precipitation before running a gel. Hope this helps a bit! ![]()
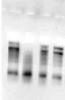
We are facing some serious problem with the DNaseI enzyme (50u/ul) (Cat. No.
EN0523)received from you few days ago. The RNA isolated from the
Plasmodium falciparum was of good quality as seen on agarose gel but after
the DNase treatment, it was getting degraded. The experiment was done twice
and same problem was encountered. In order to troubleshoot, I incubated
intact RNA (3ul; 7.5ug in each) in following reactions (Total volume of
each reaction-10ul):
1)RNA+DNase (0.5ul)- No degradation
2)RNA+DNase(0.5ul)+DNase Buffer (1ul) - Degradation
3)RNA+DNase buffer (1ul) - No degradation
4)RNA+D.W. (7ul) - No degradation
(I am attaching the gel pic of this result; Lanes are as per the reactions 1-4)
Thus, it appears that DNaseI in the DNase buffer causing some non-specific activity resulting in RNA degradation.
Please, look into the problem and get to me soon.
Thank you.